eQTL-driven tissue-specific modulation by TMPRSS2 (rs12329760) and its potential role in COVID-19 susceptibility
DOI:
https://doi.org/10.15587/2519-4798.2025.348232Keywords:
COVID-19, TMPRSS2 (rs12329760), genes, expression quantitative trait loci (eQTL), polymorphismAbstract
The aim of research – to analyze tissue-associated expression of the TMPRSS2 gene (rs12329760) using quantitative expression loci and assess its potential contribution to the formation of individual susceptibility to COVID-19.
Material and methods. Genotyping of TMPRSS2 (rs12329760) was performed for 48 patients with mild COVID-19 and 72 patients with moderate and severe disease. Peripheral blood leukocytes were used to isolate genomic DNA, and real-time polymerase chain reaction was used to genotyping gene polymorphisms. eQTL analysis was applied based on publicly available data from the QTLbase database to establish tissue-specific transcriptional effects of the TMPRSS2 variant (rs12329760).
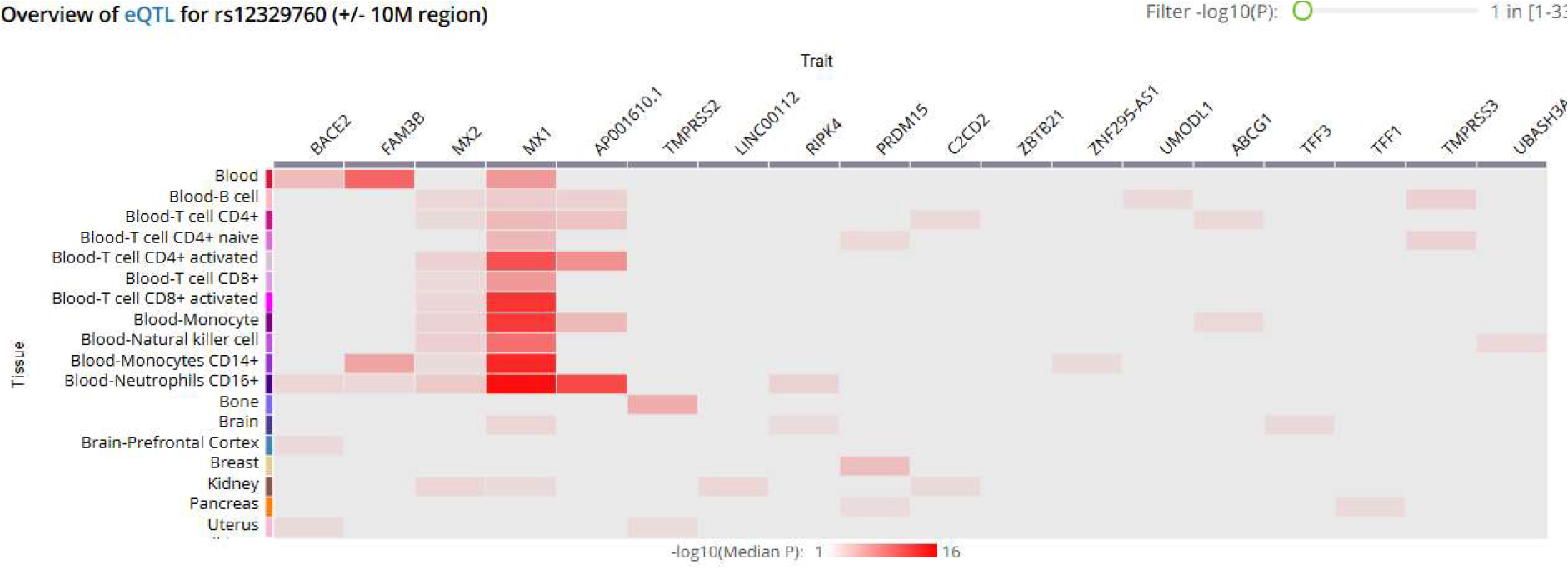
Results. It was found that TMPRSS2 (rs12329760) showed the highest number of transcripts in prostate tissue. A total of 80 eQTLs associated with rs12329760 were identified, all of which were driven exclusively by cis-regulatory mechanisms, with these associations distributed across 19 tissues and involving regulatory interactions with 18 genes. The T allele of rs12329760 was associated with significantly increased expression of MX1, MX2, and AP001610.1 in CD4⁺ T lymphocytes, circulating monocytes, and B lymphocytes. In contrast, the C allele was associated with increased expression of FAM3B and BACE2 in CD16⁺ neutrophils and CD14⁺ monocytes. Opposing regulatory effects were also found: rs12329760 was associated with transcriptional suppression of C2CD2, PRDM15, TMPRSS3, and UMODL1 in CD4⁺ T cells and B lymphocytes, predominantly mediated by the T allele.
Conclusions. The TMPRSS2 gene (rs12329760) has broad tissue-specific regulatory activity, enhancing the expression of genes involved in the formation of the human antiviral response, while involving mechanisms related to the complement system and serine protease. These transcriptional effects suggest that rs12329760 may serve as a functional modifier of susceptibility and immune response in COVID-19
References
- Chan, J. F.-W., Yuan, S., Chu, H., Sridhar, S., Yuen, K.-Y. (2024). COVID-19 drug discovery and treatment options. Nature Reviews Microbiology, 22 (7), 391–407. https://doi.org/10.1038/s41579-024-01036-y
- Sydorchuk, L., Sokolenko, M., Škoda, M., Lajcin, D., Vyklyuk, Y., Sydorchuk, R. et al. (2025). Management of Severe COVID-19 Diagnosis Using Machine Learning. Computation, 13 (10), 238. https://doi.org/10.3390/computation13100238
- Heurich, A., Hofmann-Winkler, H., Gierer, S., Liepold, T., Jahn, O., Pöhlmann, S. (2014). TMPRSS2 and ADAM17 Cleave ACE2 Differentially and Only Proteolysis by TMPRSS2 Augments Entry Driven by the Severe Acute Respiratory Syndrome Coronavirus Spike Protein. Journal of Virology, 88 (2), 1293–1307. https://doi.org/10.1128/jvi.02202-13
- Sokolenko, M., Sydorchuk, L., Sokolenko, A., Sydorchuk, R., Kamyshna, I., Sydorchuk, A. et al. (2025). Antiviral Intervention of COVID-19: Linkage of Disease Severity with Genetic Markers FGB (rs1800790), NOS3 (rs2070744) and TMPRSS2 (rs12329760). Viruses, 17 (6), 792. https://doi.org/10.3390/v17060792
- Fuentes-Prior, P. (2021). Priming of SARS-CoV-2 S protein by several membrane-bound serine proteinases could explain enhanced viral infectivity and systemic COVID-19 infection. Journal of Biological Chemistry, 296, 100135. https://doi.org/10.1074/jbc.rev120.015980
- Verma, S., Verma, S., Siddiqi, Z., Raza, S. T., Faruqui, T., Ansari, A. I. et al. (2025). Association of VDR and TMPRSS2 gene polymorphisms with COVID-19 severity: a computational and clinical study. Molecular Biology Reports, 52 (1). https://doi.org/10.1007/s11033-025-10417-2
- Paniri, A., Hosseini, M. M., Akhavan-Niaki, H. (2020). First comprehensive computational analysis of functional consequences of TMPRSS2 SNPs in susceptibility to SARS-CoV-2 among different populations. Journal of Biomolecular Structure and Dynamics, 39 (10), 3576–3593. https://doi.org/10.1080/07391102.2020.1767690
- Bizzotto, J., Sanchis, P., Abbate, M., Lage-Vickers, S., Lavignolle, R., Toro, A. et al. (2020). SARS-CoV-2 Infection Boosts MX1 Antiviral Effector in COVID-19 Patients. IScience, 23 (10), 101585. https://doi.org/10.1016/j.isci.2020.101585
- Andolfo, I., Russo, R., Lasorsa, V. A., Cantalupo, S., Rosato, B. E., Bonfiglio, F. et al. (2021). Common variants at 21q22.3 locus influence MX1 and TMPRSS2 gene expression and susceptibility to severe COVID-19. IScience, 24 (4), 102322. https://doi.org/10.1016/j.isci.2021.102322
- Ravikanth, V., Sasikala, M., Naveen, V., Latha, S. S., Parsa, K. V. L., Vijayasarathy, K. et al. (2021). A variant in TMPRSS2 is associated with decreased disease severity in COVID-19. Meta Gene, 29, 100930. https://doi.org/10.1016/j.mgene.2021.100930
- Anu, K. R., Das, S., Joseph, A. (2023). Genotype and phenotype correlations in COVID-19. Omics Approaches and Technologies in COVID-19, 41–59. https://doi.org/10.1016/b978-0-323-91794-0.00013-5
- Mir, R., Altemani, F. H., Algehainy, N. A., Alanazi, M. A., Elfaki, I., Alsayed, B. A. et al. (2025). Correction: Identification of Novel Genomic Variants in COVID-19 Patients Using Whole-Exome Sequencing: Exploring the Plausible Targets of Functional Genomics. Biochemical Genetics, 63 (6), 5001–5002. https://doi.org/10.1007/s10528-025-11024-3
- Sheehan, G., Farrell, G., Kavanagh, K. (2020). Immune priming: the secret weapon of the insect world. Virulence, 11 (1), 238–246. https://doi.org/10.1080/21505594.2020.1731137
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Maksym Sokolenko, Larysa Sydorchuk

This work is licensed under a Creative Commons Attribution 4.0 International License.
Our journal abides by the Creative Commons CC BY copyright rights and permissions for open access journals.
Authors, who are published in this journal, agree to the following conditions:
1. The authors reserve the right to authorship of the work and pass the first publication right of this work to the journal under the terms of a Creative Commons CC BY, which allows others to freely distribute the published research with the obligatory reference to the authors of the original work and the first publication of the work in this journal.
2. The authors have the right to conclude separate supplement agreements that relate to non-exclusive work distribution in the form in which it has been published by the journal (for example, to upload the work to the online storage of the journal or publish it as part of a monograph), provided that the reference to the first publication of the work in this journal is included.








