Сучасні технології дослідження геному мікобактерій
DOI:
https://doi.org/10.15587/2519-4798.2023.290596Ключові слова:
ампліфікація, геном, гібридизація, молекулярна діагностика, мікобактерії, рестрикція, секвенування, споліготипування, туберкульозАнотація
Молекулярні технології займають провідне місце в лабораторній діагностиці туберкульозу та мікобактеріозів. Успіхи досліджень генома представників роду Mycobacterium призвели до значного прогресу в розумінні еволюції, мінливості та генетичного різноманіття патогенів, а також розвитку діагностичних технологій, включаючи дослідження стійкості до протитуберкульозних препаратів.
Мета роботи: провести порівняльного дослідження спектру сучасних технологій вивчення геномів мікобактерій та їх впливу на ефективність лабораторної діагностики туберкульозу.
Матеріали та методи: пошук джерел інформації здійснювався в базах даних PubMed, Medline, Web of Science, Google Scholar. Відібрано матеріали щодо технології молекулярної діагностики туберкульозу та мікобактеріозів та визначення чутливості збудників до протитуберкульозних препаратів.
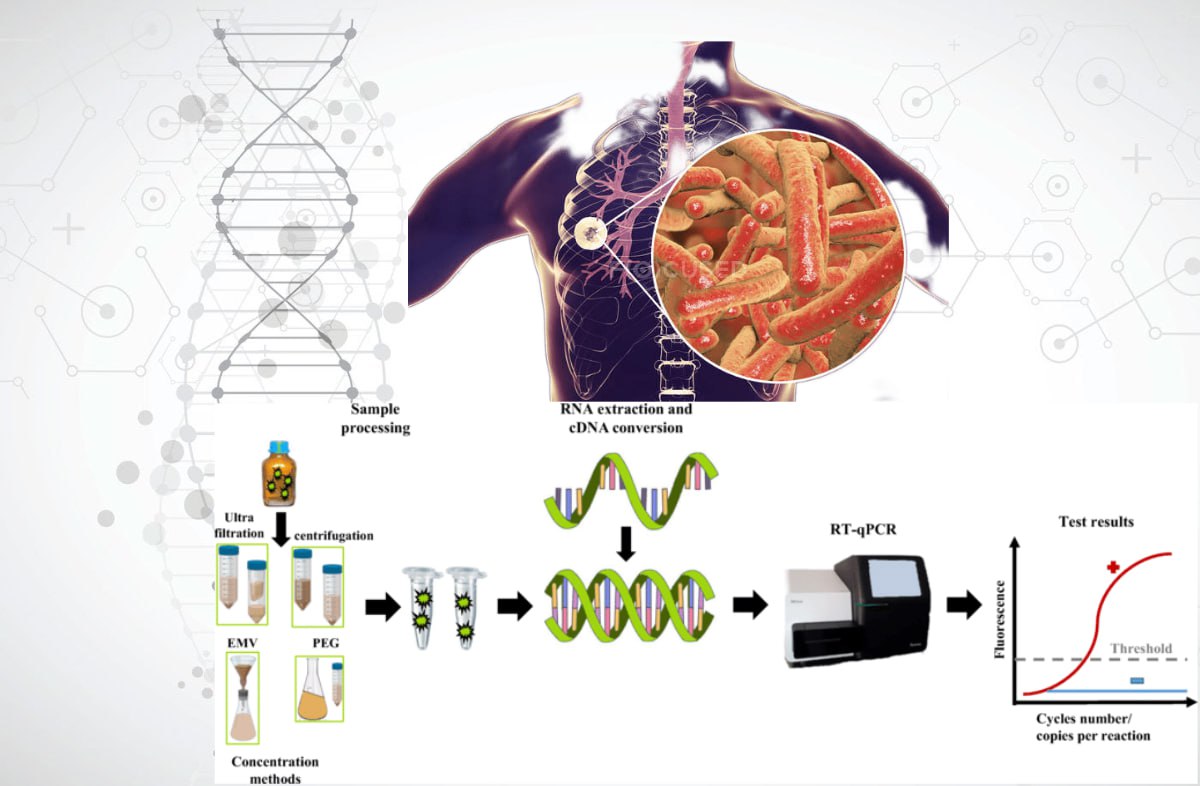
Результати: встановлено, що сучасні методи дослідження геному мікобактерій включають технології ампліфікації (ПЛР-аналіз), гібридизації, рестрикції, споліготипування, секвенування та їх різні комбінації. До основних методів належать стандартні та модифіковані протоколи ПЛР (RAPD-PCR, AP-PCR, rep-PCR, Real-time PCR, Inverse PCR, TB-LAMP, HIP, LM-PCR). Геномний рестрикційний аналіз може бути застосований при дослідженні штамів MTBC і NTM (RFLP, AFLP аналіз, генотипування MIRU-VNTR). Найефективнішим методом аналізу геному є WGS. Комплексні методи, які використовують комбінацію молекулярних технологій, дозволяють безпосередньо виявляти мікобактерії в клінічних зразках.
Висновки: широке застосування технологій дослідження геному при вивченні мікобактерій сприятиме ефективній реалізації глобальної стратегії ВООЗ щодо профілактики, лікування та боротьби з туберкульозом і мікобактеріозами
Посилання
- Global tuberculosis report 2020 (2020). World Health Organization, 208. Available at: https://apps.who.int/iris/handle/10665/336069
- Global tuberculosis report 2022 (2022). World Health Organization, 51. Available at: https://apps.who.int/iris/handle/10665/363752
- Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D. et al. (1998). Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 393 (6685), 537–544. doi: https://doi.org/10.1038/31159
- Global tuberculosis report 2019 (2019). World Health Organization, 283. Available at: https://apps.who.int/iris/handle/10665/329368
- Barbova, A. I., Zhurilo, O. A., Trofimova, P. S., Mironchenko, S. V. (2019). Experience of selection, indication and identication of non-tuberculous mycobacteria, got during I National research on study of distribution of drugresistant of tuberculosis in Ukraine. Tuberculosis, Lung Diseases, HIV Infection, 2 (37), 63–71. doi: https://doi.org/10.30978/tb2019-2-63
- Yang, S., Rothman, R. E. (2004). PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. The Lancet Infectious Diseases, 4 (6), 337–348. doi: https://doi.org/10.1016/s1473-3099(04)01044-8
- Clark, D. P., Pazdernik, N. J. (2016). DNA Synthesis In Vivo and In Vitro. Biotechnology. Applying the Genetic Revolution, 97–130. doi: https://doi.org/10.1016/b978-0-12-385015-7.00004-1
- Nurwidya, F., Handayani, D., Burhan, E., Yunus, F. (2018). Molecular Diagnosis of Tuberculosis. Chonnam Medical Journal, 54 (1). doi: https://doi.org/10.4068/cmj.2018.54.1.1
- Chia, J.-H., Wu, T.-L., Su, L.-H., Kuo, A.-J., Lai, H.-C. (2012). Direct identification of mycobacteria from smear-positive sputum samples using an improved multiplex polymerase chain reaction assay. Diagnostic Microbiology and Infectious Disease, 72 (4), 340–349. doi: https://doi.org/10.1016/j.diagmicrobio.2011.12.008
- Van Crevel, R., Hill, P. C. (2017). Tuberculosis. SECTION 2 Syndromes by Body System: The Respiratory System. Infectious Diseases, Vol. 1, 271–284. doi: https://doi.org/10.1016/b978-0-7020-6285-8.00031-9
- Percival, S. L., Williams, D. W. (2014). Mycobacterium. Microbiology of Waterborne Diseases. Microbiological Aspects and Risks, 177–207. doi: https://doi.org/10.1016/B978-0-12-415846-7.00009-3
- Shapovalova, O. V., Pozmogova, S. A., Zavgorodniy, А. І. (2022). Molecular technologies of mycobacterial research. Annals of Mechnikov Institute, 1, 9–20. doi: https://doi.org/10.5281/zenodo.7721843
- Molecular assays intended as initial tests for the diagnosis of pulmonary and extrapulmonary TB and rifampicin resistance in adults and children: rapid communication (2020). World Health Organization, 8. Available at: https://apps.who.int/iris/handle/10665/330395
- Kohli, M., Schiller, I., Dendukuri, N., Yao, M., Dheda, K., Denkinger, C. M., Schumacher, S. G., Steingart, K. R. (2021). Xpert MTB/RIF Ultra and Xpert MTB/RIF assays for extrapulmonary tuberculosis and rifampicin resistance in adults. Cochrane Database of Systematic Reviews, 2021 (1). doi: https://doi.org/10.1002/14651858.cd012768.pub3
- Tuberculosis laboratory biosafety manual (2012). World Health Organization, 50. doi: https://apps.who.int/iris/handle/10665/77949
- Pro zatverdzhennia derzhavnykh sanitarnykh norm i pravyl "Orhanizatsiia roboty laboratorii pry doslidzhenni materialu, shcho mistyt biolohichni patohenni ahenty I-IV hrup patohennosti molekuliarno-henetychnymy metodamy" (2008). Hakaz MOZU No. 26. 24.01.2008. Available at: https://zakon.rada.gov.ua/laws/show/z0088-08#Text
- Gerilovich, A. P., Stegnіi, B. T., Zavgorodnіi, A. І., Vlіzlo, V. V. et al. (2014). Molekuliarno-genetichnі metodi dіagnostiki u veterinarnіi meditcinі ta bіotekhnologіi. Kyiv: ST-Druk, 286.
- Chauhan, T. (2019). Inverse PCR: Principle, Procedure, Protocol and Applications. PCR Technology. Genetic Education. Available at: https://geneticeducation.co.in/inverse-pcr-principle-procedure-protocol-and-applications/#google_vignette
- Notomi, T., Mori, Y., Tomita, N., Kanda, H. (2015). Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. Journal of Microbiology, 53 (1), 1–5. doi: https://doi.org/10.1007/s12275-015-4656-9
- Notomi, H., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., Hase, T. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Research, 28 (12), 63e–663. doi: https://doi.org/10.1093/nar/28.12.e63
- Kim, J., Park, B. G., Lim, D. H., Jang, W. S., Nam, J., Mihn, D.-C., Lim, C. S. (2021). Development and evaluation of a multiplex loop-mediated isothermal amplification (LAMP) assay for differentiation of Mycobacterium tuberculosis and non-tuberculosis mycobacterium in clinical samples. PLOS ONE, 16 (1), e0244753. doi: https://doi.org/10.1371/journal.pone.0244753
- Molecular Test "LAMP". Available at: https://www.eiken.co.jp/en/products/lamp/
- Innovative Tool "TB LAMP". Available at: https://www.eiken.co.jp/en/ourfields/infection/tb_diagnosis/
- The use of loop-mediated isothermal amplification (TB-LAMP) for the diagnosis of pulmonary tuberculosis: policy guidance (2016). World Health Organization. Available at: https://apps.who.int/iris/handle/10665/249154
- Caulfield, A. J., Wengenack, N. L. (2016). Diagnosis of active tuberculosis disease: From microscopy to molecular techniques. Journal of Clinical Tuberculosis and Other Mycobacterial Diseases, 4, 33–43. doi: https://doi.org/10.1016/j.jctube.2016.05.005
- Line probe assays for detection of drug-resistant tuberculosis: interpretation and reporting manual for laboratory staff and clinicians (2022). World Health Organization, 31. https://apps.who.int/iris/handle/10665/354240
- Freeman, W. M., Robertson, D. J., Vrana, K. E. (2000). Fundamentals of DNA Hybridization Arrays for Gene Expression Analysis. BioTechniques, 29 (5), 1042–1055. doi: https://doi.org/10.2144/00295rv01
- Manual for selection of molecular WHO-recommended rapid diagnostic tests for detection of tuberculosis and drug-resistant tuberculosis (2022). World Health Organization, 29. https://apps.who.int/iris/handle/10665/353596
- Talbot, E. A., Raffa, B. J. (2015). Mycobacterium tuberculosis. Molecular Medical Microbiology, Vol. 3, 1637–1653. doi: https://doi.org/10.1016/B978-0-12-397169-2.00092-5
- Adam, M. A. M., Hamdan Ali, H. M., Khalil, E. A. G. (2019). Diagnostic predictive values of the hain genotype MTBDRsl assay in mycobacterial strains isolated from Sudan. Pan African Medical Journal, 32. doi: https://doi.org/10.11604/pamj.2019.32.124.12762
- Ou, X., Li, Q., Su, D., Xia, H., Wang, S., Zhao, B., Zhao, Y. (2020). A pilot study: VereMTB detection kit for rapid detection of multidrug-resistant mycobcterium tuberculosis in clinical sputum samples. PLOS ONE, 15 (3), e0228312. doi: https://doi.org/10.1371/journal.pone.0228312
- Algorithm for laboratory diagnosis and treatment-monitoring of pulmonary tuberculosis and drug-resistant tuberculosis using state-of-the-art rapid molecular diagnostic technologies: expert opinion of the European Tuberculosis Laboratory Initiative core group members for the WHO European Region (2017). World Health Organization. Regional Office for Europe. Available at: https://apps.who.int/iris/handle/10665/344108
- The use of molecular line probe assay for the detection of resistance to isoniazid and rifampicin: policy update (2016). World Health Organization, 56. https://apps.who.int/iris/handle/10665/250586
- Roychowdhury, T., Mandal, S., Bhattacharya, A. (2015). Analysis of IS6110 insertion sites provide a glimpse into genome evolution of Mycobacterium tuberculosis. Scientific Reports, 5 (1). doi: https://doi.org/10.1038/srep12567
- Jagielski, T., van Ingen, J., Rastogi, N., Dziadek, J., Mazur, P. K., Bielecki, J. (2014). Current Methods in the Molecular Typing of Mycobacterium tuberculosisand Other Mycobacteria. BioMed Research International, 2014, 1–21. doi: https://doi.org/10.1155/2014/645802
- Jagielski, T., Minias, A., van Ingen, J., Rastogi, N., Brzostek, A., Żaczek, A., Dziadek, J. (2016). Methodological and Clinical Aspects of the Molecular Epidemiology of Mycobacterium tuberculosis and Other Mycobacteria. Clinical Microbiology Reviews, 29 (2), 239–290. doi: https://doi.org/10.1128/cmr.00055-15
- Goulding, J. N., Stanley, J., Saunders, N., Arnold, C. (2000). Genome-Sequence-Based Fluorescent Amplified-Fragment Length Polymorphism Analysis of Mycobacterium tuberculosis. Journal of Clinical Microbiology, 38 (3), 1121–1126. doi: https://doi.org/10.1128/jcm.38.3.1121-1126.2000
- Kassama, Y., Shemko, M., Shetty, N., Fang, Z., MacIntire, G., Gant, V. et al. (2006). An Improved Fluorescent Amplified Fragment Length Polymorphism Method for Typing Mycobacterium tuberculosis. Journal of Clinical Microbiology, 44 (1), 288–289. doi: https://doi.org/10.1128/jcm.44.1.288-289.2006
- Varun, C. N. (2014). Clustered Regularly Interspaced Short Palindromic Repeats – Crispr. MICROBOIDS. Available at: https://varuncnmicro.blogspot.com/2014/02/clustered-regularly-interspaced-short.html
- Demay, C., Liens, B., Burguière, T., Hill, V., Couvin, D., Millet, J. et al. (2012). SITVITWEB – A publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infection, Genetics and Evolution, 12 (4), 755–766. doi: https://doi.org/10.1016/j.meegid.2012.02.004
- Brudey, K., Driscoll, J. R., Rigouts, L., Prodinger, W. M., Gori, A., Al-Hajoj, S. A. et al. (2006). Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiology, 6 (1). doi: https://doi.org/10.1186/1471-2180-6-23
- Bouakaze, C., Keyser, C., Gonzalez, A., Sougakoff, W., Veziris, N., Dabernat, H. et al. (2011). Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry-Based Single Nucleotide Polymorphism Genotyping Assay Using iPLEX Gold Technology for Identification of Mycobacterium tuberculosis Complex Species and Lineages. Journal of Clinical Microbiology, 49 (9), 3292–3299. doi: https://doi.org/10.1128/jcm.00744-11
- Burian, A. N., Zhao, W., Lo, T., Thurtle‐Schmidt, D. M. (2021). Genome sequencing guide: An introductory toolbox to whole‐genome analysis methods. Biochemistry and Molecular Biology Education, 49 (5), 815–825. doi: https://doi.org/10.1002/bmb.21561
- Bacterial Diversity Metadatabase BacDive. Available at: https://bacdive.dsmz.de/about
- The use of next-generation sequencing technologies for the detection of mutations associated with drug resistance in Mycobacterium tuberculosis complex: technical guide (2018). World Health Organization, 112. Available at: https://apps.who.int/iris/handle/10665/274443
- Zhou, X., Wu, H., Ruan, Q., Jiang, N., Chen, X., Shen, Y. et al. (2019). Clinical Evaluation of Diagnosis Efficacy of Active Mycobacterium tuberculosis Complex Infection via Metagenomic Next-Generation Sequencing of Direct Clinical Samples. Frontiers in Cellular and Infection Microbiology, 9. doi: https://doi.org/10.3389/fcimb.2019.00351
- Anochie, P. I., Onyeneke, E. C., Ogu, A. C., Onyeozirila, A. C., Aluru, S., Onyejepu, N. et al. (2012). Recent advances in the diagnosis of Mycobacterium tuberculosis. GERMS, 2 (3), 110–120. doi: https://doi.org/10.11599/germs.2012.1021
- Leão, S. C., Martin, A., Meija M, G. I., Palomino, J. C., Robledo, J., da Silva Telles, M. A., Portaels, F. (2004). Practical handbook for the phenotypic and genotypic identification of mycobacteria. Available at: http://hdl.handle.net/1854/LU-7188307
- Schlossberg, D. (Ed.) (2017). Tuberculosis and nontuberculous mycobacterial infections. Washington: ASM Press, 800. doi: https://doi.org/10.1128/9781555819866
- Schaaf, H.S., Zumla, A. (Eds.) (2009). Tuberculosis: A Comprehensive Clinical Reference. Philadelphia: Saunders Elsevier, 1046.
- Balasingham, S. V., Davidsen, T., Szpinda, I., Frye, S. A., Tønjum, T. (2009). Molecular diagnostics in tuberculosis: basis and implications for therapy. Molecular Diagnosis & Therapy, 13 (3), 137–151. doi: https://doi.org/10.1007/bf03256322
- Scarparo, C., Piccoli, P., Rigon, A., Ruggiero, G., Scagnelli, M., Piersimoni, C. (2000). Comparison of enhanced Mycobacterium tuberculosis amplified direct test with COBAS AMPLICOR Mycobacterium tuberculosis assay for direct detection of Mycobacterium tuberculosis complex in respiratory and extrapulmonary specimens. Journal of Clinical Microbiology, 38 (4), 1559–1562. doi: https://doi.org/10.1128/jcm.38.4.1559-1562.2000
- da Silva, D.A., de Pina, L.C., Rêgo, A.M., Ferreira, N.V., Redner, P., Caetano, L., Antunes, M.; Tang, Y.-W., Stratton, C. W. (Eds.) (2018). Advances in the Diagnosis of Mycobacterium tuberculosis Infection. Advanced Techniques in Diagnostic Microbiology. Vol. 2: Applications. Springer Nature Switzerland AG. 101–136. doi: https://doi.org/10.1007/978-3-319-95111-9_4
- European Centre for Disease Prevention and Control. Handbook on tuberculosis laboratory diagnostic methods in the European Union – Updated 2018 (2018). Stockholm: ECDC. Available at: https://www.ecdc.europa.eu/en/publications-data/handbook-tuberculosis-laboratory-diagnostic-methods-european-union-updated-2018
##submission.downloads##
Опубліковано
Як цитувати
Номер
Розділ
Ліцензія
Авторське право (c) 2023 Olga Shapovalova, Olena Koshova, Nataliia Filimonova

Ця робота ліцензується відповідно до Creative Commons Attribution 4.0 International License.
Наше видання використовує положення про авторські права Creative Commons CC BY для журналів відкритого доступу.
Автори, які публікуються у цьому журналі, погоджуються з наступними умовами:
1. Автори залишають за собою право на авторство своєї роботи та передають журналу право першої публікації цієї роботи на умовах ліцензії Creative Commons CC BY, котра дозволяє іншим особам вільно розповсюджувати опубліковану роботу з обов'язковим посиланням на авторів оригінальної роботи та першу публікацію роботи у цьому журналі.
2. Автори мають право укладати самостійні додаткові угоди щодо неексклюзивного розповсюдження роботи у тому вигляді, в якому вона була опублікована цим журналом (наприклад, розміщувати роботу в електронному сховищі установи або публікувати у складі монографії), за умови збереження посилання на першу публікацію роботи у цьому журналі.









