Bacterial plasmids: the role in the development of resistance to antimicrobial compounds
DOI:
https://doi.org/10.15587/2519-8025.2024.311822Keywords:
microbiology, bacteria, plasmids, genetic variability and modification, antibiotics, adaptation, resistanceAbstract
The rapid increase in the prevalence of multiple drug resistance of pathogenic microorganisms poses a critical threat to public health worldwide, which significantly contributes to the increase in patient mortality and morbidity. Classical agents, used in the past for treatment, are losing their effectiveness, moreover, many of the newer available drugs have already become targets for bacterial resistance mechanisms. As a result, the treatment of infections becomes more complicated, and the total costs of treatment increase.
Purpouse. In this work, we aimed to evaluate the role of plasmids in the development of antibiotic resistance and discuss various mechanisms of bacterial resistance to antibiotics, such as enzymatic inactivation of the antibiotic, reduction of the permeability of the outer cell membrane for the antibiotic, modification of the target mainly due to mutation, active efflux of the drug from the bacterial cell through with the help of enzymatic pumps.
Materials and methods: the search for sources of information was carried out in the databases PubMed, Medline, Web of Science, Google Scholar, as well as electronic repositories of higher education institutions and scientific institutions of Ukraine. Materials related to the research technology of genetic variability and modification of bacteria and mechanisms of resistance of microorganisms to antibiotics were selected.
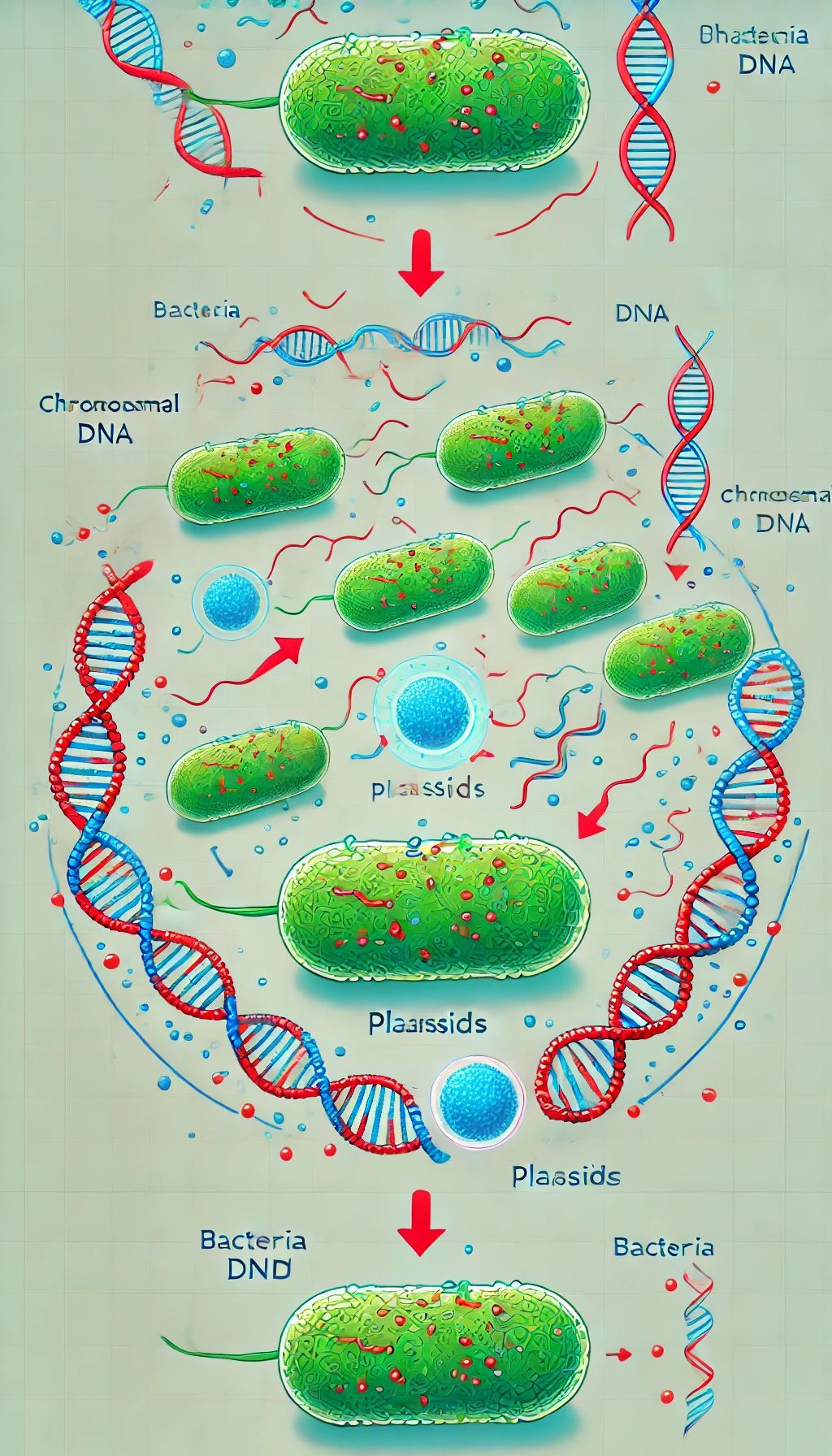
Results. As a result of evolutionary development, bacteria have acquired two separate genetic systems - chromosomal DNA and extrachromosomal, self-replicating genetic elements called plasmids.
It is bacterial plasmids that play a key role in the diffusion of specific resistance genes, in particular to antibiotics. Plasmids are non-essential parts of bacteria and are double-stranded, circular, or linear DNA molecules capable of autonomous replication, allowing bacteria to adapt to a hostile environment. Today, scientists are most interested in two properties of bacteria, caused by plasmids, - antibiotic resistance and bioremediation. The latter determines the survival and development of bacteria in various adverse conditions, including resistance to pollutants, the ability to decompose different chemical compounds, or adaptation to new ecological niches.
Conclusions. Plasmids carry genes for xenobiotic degradation and heavy metal resistance, making them useful for bioremediation of toxic chemicals in an environmentally safe manner. However, properties, such as antibiotic resistance, result from the excessive and uncontrolled use of these drugs in medicine, veterinary medicine, agriculture, and other fields. Under such conditions, there is a natural selection of those strains of pathogenic bacteria that are carriers of R-plasmids
References
- Banu, H., Prasad, K. P. (2017). Role of Plasmids in Microbiology. Journal of Aquaculture Research & Development, 8 (1). https://doi.org/10.4172/2155-9546.1000466
- Actis, L. A., Tolmasky, M. E., Crosa, J. H. (1999). Bacterial plasmids replication of extrachromosomal genetic elements encoding resistance to antimicrobial compounds. Frontiers in Bioscience, 3 (4), d43–62. https://doi.org/10.2741/a410
- Leplae, R., Hebrant, A., Wodak, S. J., Toussaint, A. (2004). ACLAME: A CLAssification of Mobile genetic Elements. Nucleic Acids Research, 32 (1), D45–D49. https://doi.org/10.1093/nar/gkh084
- Carattoli, A. (2011). Plasmids in Gram negatives: Molecular typing of resistance plasmids. International Journal of Medical Microbiology, 301 (8), 654–658. https://doi.org/10.1016/j.ijmm.2011.09.003
- Mc Ginty, S. (2012).The role of horizontal gene transfer in microbial social evolution [Doctoral dissertation, University of Zurich].
- Barlow, M. (2009). What Antimicrobial Resistance Has Taught Us About Horizontal Gene Transfer. Horizontal Gene Transfer, 397–411. https://doi.org/10.1007/978-1-60327-853-9_23
- Helinski, D. R. (2022). A Brief History of Plasmids. EcoSal Plus, 10 (1). https://doi.org/10.1128/ecosalplus.esp-0028-2021
- Hayes, W. (1968). The genetics of bacteria and their viruses. New York: John Wiley & Sons Inc.
- LEDERBERG, J., TATUM, E. L. (1946). Gene Recombination in Escherichia Coli. Nature, 158 (4016), 558–562. https://doi.org/10.1038/158558a0
- Watson, J. D., Crick, F. H. C. (1953). Molecular Structure of Nucleic Acids: A Structure for Deoxyribose Nucleic Acid. Nature, 171 (4356), 737–738. https://doi.org/10.1038/171737a0
- Kornberg, A. (1960). Biologic Synthesis of Deoxyribonucleic Acid. Science, 131 (3412), 1503–1508. https://doi.org/10.1126/science.131.3412.1503
- Watanabe, T. (1963). Infective heredity of multiple drug resistance in bacteria. Bacteriological Reviews, 27 (1), 87–115. https://doi.org/10.1128/br.27.1.87-115.1963
- Smillie, C., Garcillán-Barcia, M. P., Francia, M. V., Rocha, E. P. C., de la Cruz, F. (2010). Mobility of Plasmids. Microbiology and Molecular Biology Reviews, 74 (3), 434–452. https://doi.org/10.1128/mmbr.00020-10
- Wang, Z., Jin, L., Yuan, Z., Węgrzyn, G., Węgrzyn, A. (2009). Classification of plasmid vectors using replication origin, selection marker and promoter as criteria. Plasmid, 61 (1), 47–51. https://doi.org/10.1016/j.plasmid.2008.09.003
- Anthony, K. G., Sherburne, C., Sherburne, R., Frost, L. S. (1994). The role of the pilus in recipient cell recognition during bacterial conjugation mediated by F‐like plasmids. Molecular Microbiology, 13 (6), 939–953. https://doi.org/10.1111/j.1365-2958.1994.tb00486.x
- Zverev, V. V., Kuzmin, N. P., Zuyeva, L. A., Burova, E. I., Alexandrov, A. A., Khmel, I. A. (1984). Regions of homology in small colicinogenic plasmids. Plasmid, 12 (3), 203–205. https://doi.org/10.1016/0147-619x(84)90045-3
- Mahajan, P., Kumar, M., Bhalla, G. S., Tandel, K. (2024). Plasmid-based replicon typing: Useful tool in demonstrating the silent pandemic of plasmid-mediated multi-drug resistance in Enterobacterales. Medical Journal Armed Forces India. https://doi.org/10.1016/j.mjafi.2024.02.004
- Chun, D., Cho, D. T., Seol, S. Y., Suh, M. H., Lee, Y. C. (1984). R plasmids conferring multiple drug resistance from shigella isolated in Korea. Journal of Hygiene, 92 (2), 153–160. https://doi.org/10.1017/s0022172400064160
- Johnson, J., Warren, R. L., Branstrom, A. A. (1991). Effects of FP2 and a mercury resistance plasmid from Pseudomonas aeruginosa PA103 on exoenzyme production. Journal of Clinical Microbiology, 29 (5), 940–944. https://doi.org/10.1128/jcm.29.5.940-944.1991
- Garcillán-Barcia, M. P., Alvarado, A., de la Cruz, F. (2011). Identification of bacterial plasmids based on mobility and plasmid population biology. FEMS Microbiology Reviews, 35 (5), 936–956. https://doi.org/10.1111/j.1574-6976.2011.00291.x
- Romaniuk, L. B., Kravets, N. Y., Klymniuk, S. I., Kopcha, V. S., Dronova, O. Y. (2020). Antybiotykorezystentnist umovno-patohennykh mikroorhanizmiv: aktualnist, umovy vynyknennia, shliakhy podolannia. Infektsiini khvoroby, 4, 63–71. https://doi.org/10.11603/1681-2727.2019.4.10965
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Olena Koshova, Nataliia Filimonova, Larysa Mozghova, Iryna Tishchenko

This work is licensed under a Creative Commons Attribution 4.0 International License.
Our journal abides by the Creative Commons CC BY copyright rights and permissions for open access journals.
Authors, who are published in this journal, agree to the following conditions:
1. The authors reserve the right to authorship of the work and pass the first publication right of this work to the journal under the terms of a Creative Commons CC BY, which allows others to freely distribute the published research with the obligatory reference to the authors of the original work and the first publication of the work in this journal.
2. The authors have the right to conclude separate supplement agreements that relate to non-exclusive work distribution in the form in which it has been published by the journal (for example, to upload the work to the online storage of the journal or publish it as part of a monograph), provided that the reference to the first publication of the work in this journal is included.







