Design of non-covalent dual-acting inhibitors for proteases MPRO and PLPRO of coronavirus SARS-CoV-2 through evolutionary library generation, pharmacophore profile matching, and molecular docking calculations
DOI:
https://doi.org/10.15587/2519-4852.2024.313808Keywords:
SARS-CoV-2, Mpro protease, PLpro protease, dual inhibitors, virtual pharmacophore screening, dockingAbstract
The proteases of the SARS-CoV-2 coronavirus are crucial for the virus's life cycle, making them a prime target for developing antiviral drugs to combat COVID-19. Currently, there is a priority to develop new antiviral drugs that can target multiple viral proteins at once. In this study, we analyze the molecular mechanisms of how non-covalent inhibitors interact with the main protease (Mpro) and papain-like (PLpro) protease of SARS-CoV-2 to create a computer modelling algorithm for discovering ligands that can inhibit both Mpro and PLpro simultaneously.
Aim of the study. We aim to analyze the molecular structures involved in the interactions between current non-covalent inhibitors and the Mpro and PLpro proteases of SARS-CoV-2. The goal is to identify a common molecular structure that could be used to discover new inhibitors with a dual-acting mode using computer simulations.
Materials and Methods. LigandScout 4.5 software was used for 3D-pharmacophore analysis, virtual screening and molecular docking. AutoDock Vina 1.1.2 tools was utilized for molecular docking. Web-servers PLIP (Protein-Ligand Interaction Profiler) and Pharmit were used for studying molecular binding mechanisms. Generation of evolutionary libraries was performed by DataWarrior 6.0. Analysis and visualization were performed by Discovery Studio 2024 Suite.
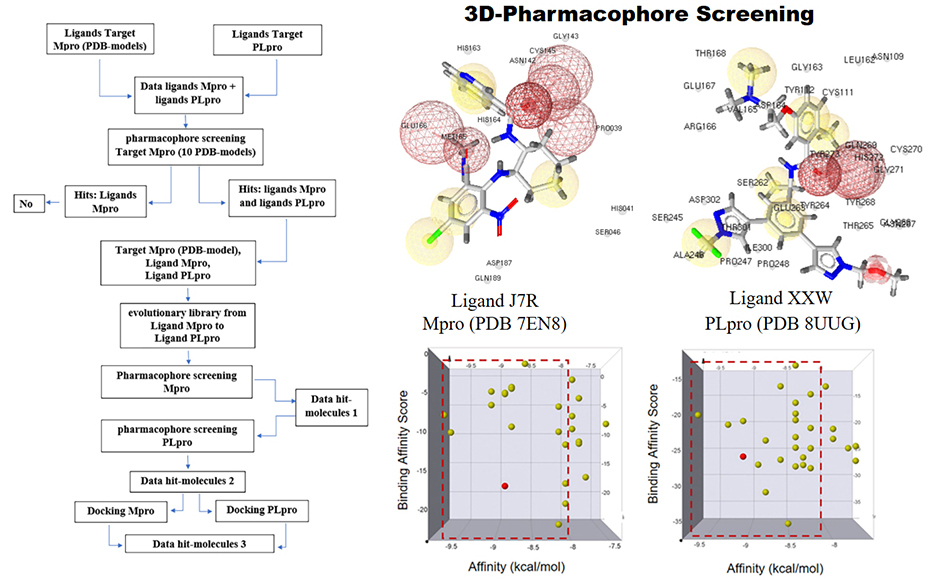
Results. Our study analyzed various models of SARS-CoV-2 protease binding sites available in the Protein Data Bank (PDB) and their corresponding non-covalent inhibitor ligands. This analysis helped identify important features of the Mpro and PLpro ligands. By comparing the pharmacophore models of Mpro ligands with the structural features of PLpro inhibitors, we identified ligands that could potentially match the binding sites of both proteases. Using the structures of these ligands, an evolutionary library was created in the DataWarrior program. Virtual screening of this library using both Mpro and PLpro pharmacophores revealed several new hit molecules. Molecular docking of these molecules into the active sites of the Mpro and PLpro proteases and calculating their binding energetics led to the identification of several molecules and their corresponding scaffolds with dual inhibition potential. These findings can be further studied in vitro with the aim of discovering drugs for COVID-19.
Conclusions. We used computer-based screening to search for ligands that could bind to both Mpro and PLpro proteases. After identifying these potential compounds, we developed synthesis methods to obtain them for further in vitro biological activity studies
Supporting Agency
- Grant No. 87/0062 (2021.01/0062) “Molecular design, synthesis and screening of new potential antiviral pharmaceutical ingredients for the treatment of infectious diseases COVID-19” from the National Research Foundation of Ukraine
References
- Yevsieieva, L. V., Lohachova, K. O., Kyrychenko, A., Kovalenko, S. M., Ivanov, V. V., Kalugin, O. N. (2023). Main and papain-like proteases as prospective targets for pharmacological treatment of coronavirus SARS-CoV-2. RSC Advances, 13 (50), 35500–35524. https://doi.org/10.1039/d3ra06479d
- Abel, R., Paredes Ramos, M., Chen, Q., Pérez-Sánchez, H., Coluzzi, F., Rocco, M. et al. (2020). Computational Prediction of Potential Inhibitors of the Main Protease of SARS-CoV-2. Frontiers in Chemistry, 8. https://doi.org/10.3389/fchem.2020.590263
- Zagórska, A., Czopek, A., Fryc, M., Jończyk, J. (2024). Inhibitors of SARS-CoV-2 Main Protease (Mpro) as Anti-Coronavirus Agents. Biomolecules, 14 (7), 797. https://doi.org/10.3390/biom14070797
- Han, H., Gracia, A. V., Røise, J. J., Boike, L., Leon, K., Schulze-Gahmen, U. et al. (2023). A covalent inhibitor targeting the papain-like protease from SARS-CoV-2 inhibits viral replication. RSC Advances, 13 (16), 10636–10641. https://doi.org/10.1039/d3ra00426k
- Prajapati, J., Patel, R., Rao, P., Saraf, M., Rawal, R., Goswami, D. (2022). Perceiving SARS-CoV-2 Mpro and PLpro dual inhibitors from pool of recognized antiviral compounds of endophytic microbes: an in silico simulation study. Structural Chemistry, 33 (5), 1619–1643. https://doi.org/10.1007/s11224-022-01932-0
- Diogo, M. A., Cabral, A. G. T., de Oliveira, R. B. (2024). Advances in the Search for SARS-CoV-2 Mpro and PLpro Inhibitors. Pathogens, 13 (10), 825. https://doi.org/10.3390/pathogens13100825
- Shi, Y., Dong, L., Ju, Z., Li, Q., Cui, Y., Liu, Y. et al. (2023). Exploring potential SARS-CoV-2 Mpro non-covalent inhibitors through docking, pharmacophore profile matching, molecular dynamic simulation, and MM-GBSA. Journal of Molecular Modeling, 29 (5). https://doi.org/10.1007/s00894-023-05534-3
- Unoh, Y., Uehara, S., Nakahara, K., Nobori, H., Yamatsu, Y., Yamamoto, S. et al. (2022). Discovery of S-217622, a Noncovalent Oral SARS-CoV-2 3CL Protease Inhibitor Clinical Candidate for Treating COVID-19. Journal of Medicinal Chemistry, 65 (9), 6499–6512. https://doi.org/10.1021/acs.jmedchem.2c00117
- Shionogi Announces Xocova® (Ensitrelvir Fumaric Acid) Obtained Standard Approval in Japan for the Treatment of SARS-CoV-2 Infection (2024). Shionogi. Available at: https://www.shionogi.com/global/en/news/2024/03/20240305.html
- Qomara, W. F., Primanissa, D. N., Amalia, S. H., Purwadi, F. V., Zakiyah, N. (2021). Effectiveness of Remdesivir, Lopinavir/Ritonavir, and Favipiravir for COVID-19 Treatment: A Systematic Review. International Journal of General Medicine, 14, 8557–8571. https://doi.org/10.2147/ijgm.s332458
- Huynh, T., Cornell, W., Luan, B. (2021). In silico Exploration of Inhibitors for SARS-CoV-2’s Papain-Like Protease. Frontiers in Chemistry, 8. https://doi.org/10.3389/fchem.2020.624163
- Sivakumar, D., Stein, M. (2021). Binding of SARS-CoV Covalent Non-Covalent Inhibitors to the SARS-CoV-2 Papain-Like Protease and Ovarian Tumor Domain Deubiquitinases. Biomolecules, 11 (6), 802. https://doi.org/10.3390/biom11060802
- Di Sarno, V., Lauro, G., Musella, S., Ciaglia, T., Vestuto, V., Sala, M. et al. (2021). Identification of a dual acting SARS-CoV-2 proteases inhibitor through in silico design and step-by-step biological characterization. European Journal of Medicinal Chemistry, 226, 113863. https://doi.org/10.1016/j.ejmech.2021.113863
- Tumskiy, R. S., Tumskaia, A. V., Klochkova, I. N., Richardson, R. J. (2023). SARS-CoV-2 proteases Mpro and PLpro: Design of inhibitors with predicted high potency and low mammalian toxicity using artificial neural networks, ligand-protein docking, molecular dynamics simulations, and ADMET calculations. Computers in Biology and Medicine, 153, 106449. https://doi.org/10.1016/j.compbiomed.2022.106449
- Kattula, B., Reddi, B., Jangam, A., Naik, L., Adimoolam, B. M., Vavilapalli, S. et al. (2023). Development of 2-chloroquinoline based heterocyclic frameworks as dual inhibitors of SARS-CoV-2 MPro and PLPro. International Journal of Biological Macromolecules, 242, 124772. https://doi.org/10.1016/j.ijbiomac.2023.124772
- Kyrychenko, A., Bylov, I., Geleverya, A., Kovalenko, S., Zhuravel, I., Fetyukhin, V., Langer, T. (2024). Computer-aided rational design and synthesis of new potential antihypertensive agents among 1,2,3-triazole-containing nifedipine analogs. ScienceRise: Pharmaceutical Science, 3 (49), 4–12. https://doi.org/10.15587/2519-4852.2024.291626
- Lohachova, K. O., Sviatenko, A. S., Kyrychenko, A., Ivanov, V. V., Langer, T., Kovalenko, S. M., Kalugin, O. N. (2024). Computer-aided drug design of novel nirmatrelvir analogs inhibiting main protease of Coronavirus SARS-CoV-2. Journal of Applied Pharmaceutical Science, 14 (5), 232–239. https://doi.org/10.7324/japs.2024.158114
- Shen, J.-X., Du, W.-W., Xia, Y.-L., Zhang, Z.-B., Yu, Z.-F., Fu, Y.-X., Liu, S.-Q. (2023). Identification of and Mechanistic Insights into SARS-CoV-2 Main Protease Non-Covalent Inhibitors: An In-Silico Study. International Journal of Molecular Sciences, 24 (4), 4237. https://doi.org/10.3390/ijms24044237
- Wolber, G., Langer, T. (2004). LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. Journal of Chemical Information and Modeling, 45 (1), 160–169. https://doi.org/10.1021/ci049885e
- Lipinski, C. A., Lombardo, F., Dominy, B. W., Feeney, P. J. (2012). Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Advanced Drug Delivery Reviews, 64, 4–17. https://doi.org/10.1016/j.addr.2012.09.019
- Sander, T., Freyss, J., von Korff, M., Rufener, C. (2015). DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. Journal of Chemical Information and Modeling, 55 (2), 460–473. https://doi.org/10.1021/ci500588j
- RCSB Protein Data Bank. Available at: https://www.rcsb.org
- Goodsell, D. S., Morris, G. M., Olson, A. J. (1996). Automated docking of flexible ligands: Applications of autodock. Journal of Molecular Recognition, 9(1), 1–5. https://doi.org/10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6
- Trott, O., Olson, A. J. (2009). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31 (2), 455–461. https://doi.org/10.1002/jcc.21334
- Protein-Ligand Interaction Profiler. Available at: https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index
- Pharmit. Available at: https://pharmit.csb.pitt.edu
- Adasme, M. F., Linnemann, K. L., Bolz, S. N., Kaiser, F., Salentin, S., Haupt, V. J., Schroeder, M. (2021). PLIP 2021: expanding the scope of the protein–ligand interaction profiler to DNA and RNA. Nucleic Acids Research, 49 (W1), W530–W534. https://doi.org/10.1093/nar/gkab294
- Sunseri, J., Koes, D. R. (2016). Pharmit: interactive exploration of chemical space. Nucleic Acids Research, 44 (W1), W442–W448. https://doi.org/10.1093/nar/gkw287
- Anokhin, D., Kovalenko, S., Trostianko, P., Kyrychenko, A., Zakharov, A., Zubatiuk, T. et al. (2024). Towards the discovery of molecules with anti-COVID-19 activity: Relationships between screening and docking results. Kharkiv University Bulletin. Chemical Series, 42, 6–14. https://doi.org/10.26565/2220-637X-2024-42-01
- Silin, O. V., Savchenko, T. I., Kovalenko, S. M., Nikitchenko, V. M., Ivachtchenko, A. V. (2004). Synthesis of Novel 5H-Pyrazolo[4,3-c]quinolines. Heterocycles, 63 (8), 1883–1890. https://doi.org/10.3987/com-04-10092
- Savchenko, T. I., Silin, O. V., Kovalenko, S. M., Musatov, V. I., Nikitchenko, V. M., Ivachtchenko, A. V. (2007). Alkylation of 3‐Phenyl‐1H‐pyrazolo[4,3‐c] quinoline: Theoretical Analysis of Regioselectivity. Synthetic Communications, 37 (8), 1321–1330. https://doi.org/10.1080/00397910701227077
- Xiao, Y.-Q., Long, J., Zhang, S.-S., Zhu, Y.-Y., Gu, S.-X. (2024). Non-peptidic inhibitors targeting SARS-CoV-2 main protease: A review. Bioorganic Chemistry, 147, 107380. https://doi.org/10.1016/j.bioorg.2024.107380
- Yang, Y., Luo, Y.-D., Zhang, C.-B., Xiang, Y., Bai, X.-Y., Zhang, D., Fu, Z.-Y. et al. (2024). Progress in Research on Inhibitors Targeting SARS-CoV-2 Main Protease (Mpro). ACS Omega, 9 (32), 34196–34219. https://doi.org/10.1021/acsomega.4c03023
- Tyndall, J. D. A. (2022). S-217622, a 3CL Protease Inhibitor and Clinical Candidate for SARS-CoV-2. Journal of Medicinal Chemistry, 65 (9), 6496–6498. https://doi.org/10.1021/acs.jmedchem.2c00624
- Kawajiri, T., Kijima, A., Iimuro, A., Ohashi, E., Yamakawa, K., Agura, K. et al. (2023). Development of a Manufacturing Process toward the Convergent Synthesis of the COVID-19 Antiviral Ensitrelvir. ACS Central Science, 9 (4), 836–843. https://doi.org/10.1021/acscentsci.2c01203
- Song, L., Gao, S., Ye, B., Yang, M., Cheng, Y., Kang, D. et al. (2024). Medicinal chemistry strategies towards the development of non-covalent SARS-CoV-2 Mpro inhibitors. Acta Pharmaceutica Sinica B, 14 (1), 87–109. https://doi.org/10.1016/j.apsb.2023.08.004
- Taylor, A. J., Amporndanai, K., Rietz, T. A., Zhao, B., Thiruvaipati, A., Wei, Q. et al. (2024). Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PLpro). ACS Medicinal Chemistry Letters, 15 (8), 1351–1357. https://doi.org/10.1021/acsmedchemlett.4c00238
- Magwaza, N. N., Mushebenge, A. G.-A., Ugbaja, S. C., Mbatha, N. A., Khan, R. B., Kumalo, H. M. (2024). Mechanistic Insights into Targeting SARS-CoV-2 Papain-like Protease in the Evolution and Management of COVID-19. BioChem, 4 (3), 268–299. https://doi.org/10.3390/biochem4030014
- Schimunek, J., Seidl, P., Elez, K., Hempel, T., Le, T., Noé, F. et al. (2023). A community effort in SARS‐CoV‐2 drug discovery. Molecular Informatics, 43 (1). https://doi.org/10.1002/minf.202300262
- Ivanov, V., Lohachova, K., Kolesnik, Y., Zakharov, A., Yevsieieva, L., Kyrychenko, A. et al. (2023). Recent advances in computational drug discovery for therapy against coronavirus SARS-CoV-2. ScienceRise: Pharmaceutical Science, 6 (46), 4–24. https://doi.org/10.15587/2519-4852.2023.290318
- Puhl, A. C., Godoy, A. S., Noske, G. D., Nakamura, A. M., Gawriljuk, V. O., Fernandes, R. S. et al. (2023). Discovery of PLpro and Mpro Inhibitors for SARS-CoV-2. ACS Omega, 8 (25), 22603–22612. https://doi.org/10.1021/acsomega.3c01110
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Larysa Yevsieieva, Pavlo Trostianko, Alexander Kyrychenko, Volodymyr Ivanov, Sergiy Kovalenko, Oleg Kalugin

This work is licensed under a Creative Commons Attribution 4.0 International License.
Our journal abides by the Creative Commons CC BY copyright rights and permissions for open access journals.






