A comparison study of artificial intelligence-driven no-code applications for drug discovery and development
DOI:
https://doi.org/10.15587/2519-4852.2024.318920Keywords:
AI-driven applications, drug discovery, no-code platforms, machine learning, pharmaceutical researchAbstract
The aim. The aim of this study was to evaluate the functionality and effectiveness of selected AI-driven no-code applications in drug discovery. This research assessed ease of use, interface design, user experience, speed, resource utilisation, accuracy, and scalability to determine their suitability for various drug development tasks.
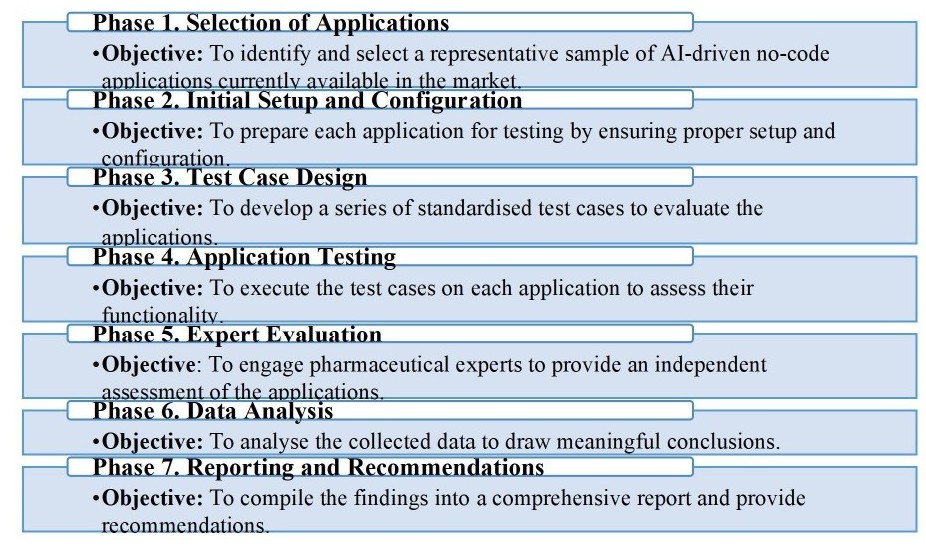
Materials and methods. The study used an evaluation methodology to test six AI-driven no-code applications: Insilico Medicine's Pharma.AI, Atomwise, Schrödinger's LiveDesign, Exscientia, BenevolentAI, and Cyclica. Quantitative data were collected from performance metrics, and qualitative data were obtained through expert interviews. Data analysis was conducted using descriptive statistics, repeated measures ANOVA, and post hoc Tukey's Honestly Significant Difference (HSD) tests.
Results.The analysis revealed that Insilico Medicine's Pharma.AI and Atomwise consistently outperformed other applications regarding usability and predictive accuracy. Schrödinger's LiveDesign demonstrated high accuracy but required significant computational resources. BenevolentAI and Exscientia showed limitations in usability and accuracy, particularly in toxicity prediction. Cyclica was noted for its ease of use but was less effective in scalability and resource utilisation.
Conclusions. The findings provide valuable insights for researchers and pharmaceutical companies, guiding the integration and application of AI-driven solutions to accelerate the drug discovery process and improve the success rate of developing new therapeutic drugs. Future research should focus on broadening the evaluation to include more diverse scenarios and real-world applications to further validate and enhance these tools
References
- Austin, D., Hayford, T. (2021). Research and development in the pharmaceutical industry. CBO. Available at: https://www.cbo.gov/publication/57126
- Schuhmacher, A., Hinder, M., von Stegmann und Stein, A., Hartl, D., Gassmann, O. (2023). Analysis of pharma R&D productivity – a new perspective needed. Drug Discovery Today, 28 (10), 103726. https://doi.org/10.1016/j.drudis.2023.103726
- Blanco-González, A., Cabezón, A., Seco-González, A., Conde-Torres, D., Antelo-Riveiro, P., Piñeiro, Á., Garcia-Fandino, R. (2023). The Role of AI in Drug Discovery: Challenges, Opportunities, and Strategies. Pharmaceuticals, 16 (6), 891. https://doi.org/10.3390/ph16060891
- Paul, D., Sanap, G., Shenoy, S., Kalyane, D., Kalia, K., Tekade, R. K. (2021). Artificial intelligence in drug discovery and development. Drug Discovery Today, 26 (1), 80–93. https://doi.org/10.1016/j.drudis.2020.10.010
- Xu, Y., Liu, X., Cao, X., Huang, C., Liu, E., Qian, S. et al. (2021). Artificial intelligence: A powerful paradigm for scientific research. The Innovation, 2 (4), 100179. https://doi.org/10.1016/j.xinn.2021.100179
- Singh, S., Kumar, R., Payra, S., Singh, S. K. (2023). Artificial Intelligence and Machine Learning in Pharmacological Research: Bridging the Gap Between Data and Drug Discovery. Cureus. https://doi.org/10.7759/cureus.44359
- Kim, H., Kim, E., Lee, I., Bae, B., Park, M., Nam, H. (2020). Artificial Intelligence in Drug Discovery: A Comprehensive Review of Data-driven and Machine Learning Approaches. Biotechnology and Bioprocess Engineering, 25 (6), 895–930. https://doi.org/10.1007/s12257-020-0049-y
- Ghislat, G., Hernandez-Hernandez, S., Piyawajanusorn, C., Ballester, P. J. (2024). Data-centric challenges with the application and adoption of artificial intelligence for drug discovery. Expert Opinion on Drug Discovery, 19(11), 1297–1307. https://doi.org/10.1080/17460441.2024.2403639
- Kolluri, S., Lin, J., Liu, R., Zhang, Y., Zhang, W. (2022). Machine Learning and Artificial Intelligence in Pharmaceutical Research and Development: a Review. The AAPS Journal, 24 (1). https://doi.org/10.1208/s12248-021-00644-3
- Lavecchia, A. (2015). Machine-learning approaches in drug discovery: methods and applications. Drug Discovery Today, 20 (3), 318–331. https://doi.org/10.1016/j.drudis.2014.10.012
- Gomes, J., Ramsundar, B., Feinberg, E. N., Pande, V. S. (2017). Atomic convolutional networks for predicting protein-ligand binding affinity. https://doi.org/10.48550/arXiv.1703.10603
- Zhavoronkov, A., Ivanenkov, Y. A., Aliper, A., Veselov, M. S., Aladinskiy, V. A., Aladinskaya, A. V. et al. (2019). Deep learning enables rapid identification of potent DDR1 kinase inhibitors. Nature Biotechnology, 37 (9), 1038–1040. https://doi.org/10.1038/s41587-019-0224-x
- Stokes, J. M., Yang, K., Swanson, K., Jin, W., Cubillos-Ruiz, A., Donghia, N. M. et al. (2020). A deep learning approach to antibiotic discovery. Cell, 181 (2), 475–483. https://doi.org/10.1016/j.cell.2020.01.021
- Vamathevan, J., Clark, D., Czodrowski, P., Dunham, I., Ferran, E., Lee, G. et al. (2019). Applications of machine learning in drug discovery and development. Nature Reviews Drug Discovery, 18 (6), 463–477. https://doi.org/10.1038/s41573-019-0024-5
- Libbrecht, M. W., Noble, W. S. (2015). Machine learning applications in genetics and genomics. Nature Reviews Genetics, 16 (6), 321–332. https://doi.org/10.1038/nrg3920
- Zhou, J., Troyanskaya, O. G. (2015). Predicting effects of noncoding variants with deep learning–based sequence model. Nature Methods, 12 (10), 931–934. https://doi.org/10.1038/nmeth.3547
- Tiwary, S., Levy, R., Gutenbrunner, P., Salinas Soto, F., Palaniappan, K. K., Deming, L. et al. (2019). High-quality MS/MS spectrum prediction for data-dependent and data-independent acquisition data analysis. Nature Methods, 16 (6), 519–525. https://doi.org/10.1038/s41592-019-0427-6
- Casadio, R., Martelli, P. L., Savojardo, C. (2022). Machine learning solutions for predicting protein–protein interactions. WIREs Computational Molecular Science, 12(6). https://doi.org/10.1002/wcms.1618
- Kourou, K., Exarchos, T. P., Exarchos, K. P., Karamouzis, M. V., Fotiadis, D. I. (2015). Machine learning applications in cancer prognosis and prediction. Computational and Structural Biotechnology Journal, 13, 8–17. https://doi.org/10.1016/j.csbj.2014.11.005
- Esteva, A., Kuprel, B., Novoa, R. A., Ko, J., Swetter, S. M., Blau, H. M., Thrun, S. (2017). Dermatologist-level classification of skin cancer with deep neural networks. Nature, 542 (7639), 115–118. https://doi.org/10.1038/nature21056
- Chen, H., Engkvist, O., Wang, Y., Olivecrona, M., Blaschke, T. (2018). The rise of deep learning in drug discovery. Drug Discovery Today, 23 (6), 1241–1250. https://doi.org/10.1016/j.drudis.2018.01.039
- Wallach, I., Dzamba, M., Heifets, A. (2015). AtomNet: A deep convolutional neural network for bioactivity prediction in structure-based drug discovery. arXiv preprint arXiv:1510.02855. https://doi.org/10.48550/arXiv.1510.02855
- Xu, Y., Dai, Z., Chen, F., Gao, S., Pei, J., Lai, L. (2015). Deep Learning for Drug-Induced Liver Injury. Journal of Chemical Information and Modeling, 55 (10), 2085–2093. https://doi.org/10.1021/acs.jcim.5b00238
- Klambauer, G., Unterthiner, T., Mayr, A., Hochreiter, S. (2017). DeepTox: Toxicity prediction using deep learning. Toxicology Letters, 280, S69. https://doi.org/10.1016/j.toxlet.2017.07.175
- Cavasotto, C. N., Scardino, V. (2022). Machine Learning Toxicity Prediction: Latest Advances by Toxicity End Point. ACS Omega, 7(51), 47536–47546. https://doi.org/10.1021/acsomega.2c05693
- Nam, S., Kim, D., Jung, W., Zhu, Y. (2022). Understanding the Research Landscape of Deep Learning in Biomedical Science: Scientometric Analysis. Journal of Medical Internet Research, 24 (4), e28114. https://doi.org/10.2196/28114
- Kavalci, E., Hartshorn, A. (2023). Improving clinical trial design using interpretable machine learning based prediction of early trial termination. Scientific Reports, 13 (1). https://doi.org/10.1038/s41598-023-27416-7
- Esteva, A., Chou, K., Yeung, S., Naik, N., Madani, A., Mottaghi, A., Liu, Y., Topol, E., Dean, J., Socher, R. (2021). Deep learning-enabled medical computer vision. Npj Digital Medicine, 4 (1). https://doi.org/10.1038/s41746-020-00376-2
- Ghassemi, M., Naumann, T., Schulam, P., Beam, A. L., Chen, I. Y., Ranganath, R. (2020). A Review of Challenges and Opportunities in Machine Learning for Health. AMIA Joint Summits on Translational Science proceedings, 191–200. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7233077/pdf/3268580.pdf
- Ghassemi, M., Naumann, T., Schulam, P., Beam, A. L., Chen, I. Y., Ranganath, R. (2019). Practical guidance on artificial intelligence for health-care data. The Lancet Digital Health, 1 (4), e157–e159. https://doi.org/10.1016/s2589-7500(19)30084-6
- Ghosh, S., Das, N., Chakraborty, S., Chakrabarti, A. (2020). Understanding deep learning techniques for image segmentation. arXivLabs. https://doi.org/10.48550/arXiv.1907.06119
- Obermeyer, Z., Powers, B., Vogeli, C., Mullainathan, S. (2019). Dissecting racial bias in an algorithm used to manage the health of populations. Science, 366 (6464), 447–453. https://doi.org/10.1126/science.aax2342
- Jiménez-Luna, J., Grisoni, F., Weskamp, N., Schneider, G. (2021). Artificial intelligence in drug discovery: recent advances and future perspectives. Expert Opinion on Drug Discovery, 16 (9), 949–959. https://doi.org/10.1080/17460441.2021.1909567
- Lundberg, S. M., Lee, S. I. (2017). A unified approach to interpreting model predictions. Proceedings of the 31st International Conference on Neural Information Processing Systems, 4768–4777. https://doi.org/10.48550/arXiv.1705.07874
- Ribeiro, M. T., Singh, S., Guestrin, C. (2016). “Why should I trust you?” Explaining the predictions of any classifier. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 1135–1144. https://doi.org/10.1145/2939672.2939778
- Bauer, C. (2020). Multi-Method Evaluation: Leveraging Multiple Methods to Answer What You Were Looking For. Proceedings of the 2020 Conference on Human Information Interaction and Retrieval (CHIIR '20). Association for Computing Machinery, 472–474. https://doi.org/10.1145/3343413.3378015
- Pharma.AI. Insilico Medicine’s. Available at: https://pharma.ai/
- Atomwise. Available at: https://www.atomwise.com/
- Schrödinger’s. LiveDesign. Available at: https://www.schrodinger.com/
- Exscientia. Available at: https://www.exscientia.com/
- Benevolent AI. Available at: https://www.benevolent.com//
- Cyclica. Available at: https://cyclicarx.com/?lang=en
- The jamovi project (2024). jamovi (Version 2.5) [Computer Software]. Available at: https://www.jamovi.org/
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Iryna Nizhenkovska, Tetyana Reva, Olena Kuznetsova, Oleksii Nizhenkovskyi, Oksana Chkhalo

This work is licensed under a Creative Commons Attribution 4.0 International License.
Our journal abides by the Creative Commons CC BY copyright rights and permissions for open access journals.






