Цілеспрямований структурний дизайн молекулярних скаффолдів для дуальних пептидоміметичних інгібіторів протеаз SARS-CoV-2 MPRO та PLPRO
DOI:
https://doi.org/10.15587/2519-4852.2025.337951Ключові слова:
SARS-CoV-2, протеаза Mpro, протеаза PLpro, пептидоміметики, дуальні інгібітори, молекулярний докінгАнотація
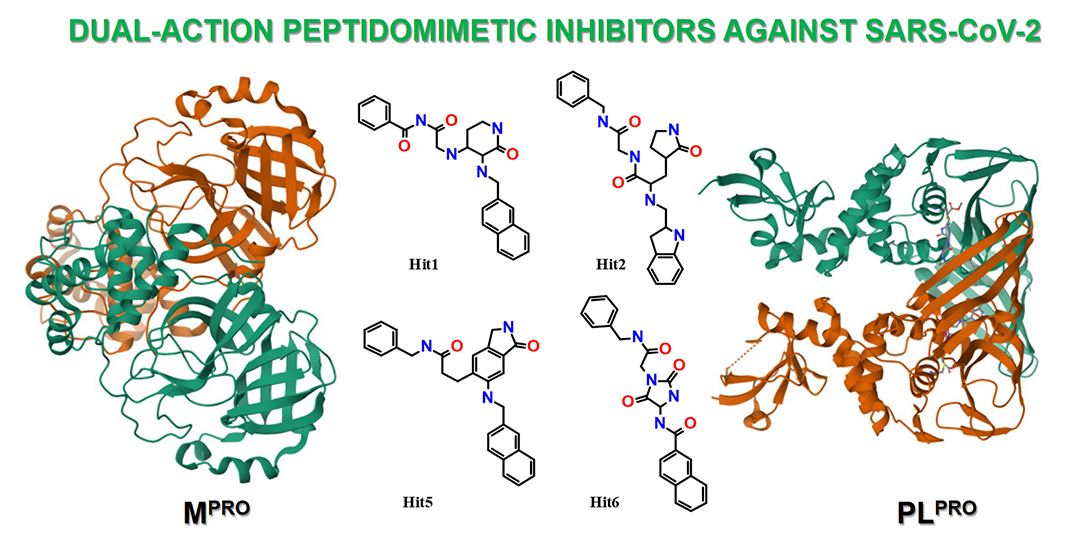
Дві протеази SARS-CoV-2 коронавірусу – головна протеаза (Mpro або 3CLpro) та папаїн-подібна протеаза (PLpro) – є ключовими ферментами, необхідними для успішної реплікації вірусу в клітинах. Обидві протеази стали головними мішенями для розробки лікарських засобів проти SARS-CoV-2. Перспектива досягнення подвійного інгібуючого ефекту викликає значний інтерес до створення подвійних інгібіторів, як комплексних терапевтичних засобів проти SARS-CoV-2. У цій статті ми описуємо розробку і оцінку in silico серії нових пептидоміметичних молекул, розроблених як інгібітори подвійної дії Mpro та PLpro SARS-CoV-2, та їх синтез. Ми застосували комбінований підхід, який розпочався з розробки базової молекулярної моделі, що враховувала субстратну специфічність активного центру кожної протеази. Раціональний дизайн in silico привів до створення ряду пептидоміметиків. Подальший аналіз зв'язування цих сполук з активними центрами обох протеаз дозволив ідентифікувати низку нових структурних хітів, зокрема гідантоїнових похідних, як потенційних подвійних інгібіторів Mpro та PLpro.
Мета дослідження. Це дослідження спрямоване на визначення спільного молекулярного каркасу для структурного дизайну інгібіторів подвійної дії проти протеаз SARS-CoV-2 Mpro та PLpro. Дослідження включає рецептор-орієнтований молекулярний докінг, оптимізацію in silico, вибір та синтез найбільш активних структур-кандидатів для подальших експериментальних досліджень in vitro.
Матеріали та методи. Програмне забезпечення LigandScout 4.5 для 3D-фармакофорного аналізу, візуалізації та молекулярного докінгу, інструменти AutoDock Vina 1.1 для молекулярного докінгу. Веб-сервери PLIP (Protein-Ligand Interaction Profiler) для вивчення механізмів молекулярного зв'язування. Програмне забезпечення DataWarrior 6.0 для створення бібліотеки молекулярних структур, розрахунків фізико-хімічних властивостей та аналізу молекулярних каркасів. Веб-сервери SwissADME для прогнозування параметрів ADME, фармакокінетичних властивостей малих молекул у якості лікарських засобів.
Результати. Аналіз субстратної специфічності сайтів зв'язування протеаз Mpro і PLpro дозволив ідентифікувати спільну амінокислотну послідовність, яка включає загальні елементи розпізнавання для кожної протеази. Шляхом раціональній модифікації функціональних груп у цій початковій базовій структурі, використовуючи принцип ізостеричної заміни та застосовуючи некласичні біоізостери, ми розробили серію пептидоміметичних каркасів. Молекулярний докінг, проведений в активних центрах як Mpro, так і PLpro, у поєднанні з оцінкою відповідних показників енергії зв'язування (ккал/моль), виявив кілька структур, що демонструють потенціал для подвійного інгібування. Серед них похідні гідантоїну продемонстрували найсильнішу спорідненість зв'язування з активними центрами обох протеаз.
Висновки. Визначено перспективні базові пептидоміметичні молекулярні структури, що показали in silico подвійну інгібуючу активність проти протеаз SARS-CoV-2. Нами ідентифіковано новий клас гідантоїнових похідних як інгібіторів як Mpro, так і PLpro SARS-CoV-2. Розроблені методи синтезу дозволили успішно отримати ці сполуки для подальших досліджень in vitro
Спонсор дослідження
- Grant No. 96/0062 (2021.01/0062) “Molecular design, synthesis and screening of new potential antiviral pharmaceutical ingredients for the treatment of infectious diseases COVID-19” from the National Research Foundation of Ukraine
Посилання
- Yevsieieva, L. V., Lohachova, K. O., Kyrychenko, A., Kovalenko, S. M., Ivanov, V. V., Kalugin, O. N. (2023). Main and papain-like proteases as prospective targets for pharmacological treatment of coronavirus SARS-CoV-2. RSC Advances, 13 (50), 35500–35524. https://doi.org/10.1039/d3ra06479d
- Capasso, C., Nocentini, A., Supuran, C. T. (2020). Protease inhibitors targeting the main protease and papain-like protease of coronaviruses. Expert Opinion on Therapeutic Patents, 31 (4), 309–324. https://doi.org/10.1080/13543776.2021.1857726
- Wang, Y., Xu, B., Ma, S., Wang, H., Shang, L., Zhu, C., Ye, S. (2022). Discovery of SARS-CoV-2 3CLPro Peptidomimetic Inhibitors through the Catalytic Dyad Histidine-Specific Protein-Ligand Interactions. International Journal of Molecular Sciences, 23 (4), 2392. https://doi.org/10.3390/ijms23042392
- Kerti, L., Frecer, V. (2024). Design of inhibitors of SARS-CoV-2 papain-like protease deriving from GRL0617: Structure–activity relationships. Bioorganic & Medicinal Chemistry, 113, 117909. https://doi.org/10.1016/j.bmc.2024.117909
- Lohachova, K. O., Kyrychenko, A., Kalugin, O. N. (2025). Benchmarking biomolecular force fields for molecular dynamics simulations of native fold and enzymatic activity of SARS-CoV-2 papain-like protease. Heliyon, 11 (12), e43578. https://doi.org/10.1016/j.heliyon.2025.e43578
- Garnsey, M. R., Robinson, M. C., Nguyen, L. T., Cardin, R., Tillotson, J., Mashalidis, E. et al. (2024). Discovery of SARS-CoV-2 papain-like protease (PLpro) inhibitors with efficacy in a murine infection model. bioRxiv. https://doi.org/10.1101/2024.01.26.577395
- Yevsieieva, L., Trostianko, P., Kyrychenko, A., Ivanov, V., Kovalenko, S., Kalugin, O. (2024). Design of non-covalent dual-acting inhibitors for proteases MPRO and PLPRO of coronavirus SARS-CoV-2 through evolutionary library generation, pharmacophore profile matching, and molecular docking calculations. ScienceRise: Pharmaceutical Science, 6 (52), 15–26. https://doi.org/10.15587/2519-4852.2024.313808
- Ma, C., Sacco, M. D., Xia, Z., Lambrinidis, G., Townsend, J. A., Hu, Y. et al. (2021). Discovery of SARS-CoV-2 Papain-like Protease Inhibitors through a Combination of High-Throughput Screening and a FlipGFP-Based Reporter Assay. ACS Central Science, 7 (7), 1245–1260. https://doi.org/10.1021/acscentsci.1c00519
- Lucas, S. C. C., Blackwell, J. H., Hewitt, S. H., Semple, H., Whitehurst, B. C., Xu, H. (2024). Covalent hits and where to find them. SLAS Discovery, 29 (3), 100142. https://doi.org/10.1016/j.slasd.2024.01.003
- Ivanov, V., Lohachova, K., Kolesnik, Y., Zakharov, A., Yevsieieva, L., Kyrychenko, A. et al. (2023). Recent advances in computational drug discovery for therapy against coronavirus SARS-CoV-2. ScienceRise: Pharmaceutical Science, 6 (46), 4–24. https://doi.org/10.15587/2519-4852.2023.290318
- Zhang, L., Lin, D., Sun, X., Curth, U., Drosten, C., Sauerhering, L. et al. (2020). Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science, 368 (6489), 409–412. https://doi.org/10.1126/science.abb3405
- Xia, Z., Sacco, M., Hu, Y., Ma, C., Meng, X., Zhang, F. et al. (2021). Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir. ACS Pharmacology & Translational Science, 4 (4), 1408–1421. https://doi.org/10.1021/acsptsci.1c00099
- Citarella, A., Scala, A., Piperno, A., Micale, N. (2021). SARS-CoV-2 Mpro: A Potential Target for Peptidomimetics and Small-Molecule Inhibitors. Biomolecules, 11 (4), 607. https://doi.org/10.3390/biom11040607
- Bege, M., Borbás, A. (2024). The Design, Synthesis and Mechanism of Action of Paxlovid, a Protease Inhibitor Drug Combination for the Treatment of COVID-19. Pharmaceutics, 16 (2), 217. https://doi.org/10.3390/pharmaceutics16020217
- Lohachova, K. O., Sviatenko, A. S., Kyrychenko, A., Ivanov, V. V., Langer, T., Kovalenko, S. M., Kalugin, O. N. (2024). Computer-aided drug design of novel nirmatrelvir analogs inhibiting main protease of Coronavirus SARS-CoV-2. Journal of Applied Pharmaceutical Science, 14 (5), 232–239. https://doi.org/10.7324/japs.2024.158114
- Arafet, K., Serrano-Aparicio, N., Lodola, A., Mulholland, A. J., González, F. V., Świderek, K., Moliner, V. (2021). Mechanism of inhibition of SARS-CoV-2 Mpro by N3 peptidyl Michael acceptor explained by QM/MM simulations and design of new derivatives with tunable chemical reactivity. Chemical Science, 12 (4), 1433–1444. https://doi.org/10.1039/d0sc06195f
- Bauer, R. A. (2015). Covalent inhibitors in drug discovery: from accidental discoveries to avoided liabilities and designed therapies. Drug Discovery Today, 20 (9), 1061–1073. https://doi.org/10.1016/j.drudis.2015.05.005
- Singh, J., Petter, R. C., Baillie, T. A., Whitty, A. (2011). The resurgence of covalent drugs. Nature Reviews Drug Discovery, 10 (4), 307–317. https://doi.org/10.1038/nrd3410
- Schaefer, D., Cheng, X. (2023). Recent Advances in Covalent Drug Discovery. Pharmaceuticals, 16 (5), 663. https://doi.org/10.3390/ph16050663
- Hu, Y., Ma, C., Szeto, T., Hurst, B., Tarbet, B., Wang, J. (2021). Boceprevir, Calpain Inhibitors II and XII, and GC-376 Have Broad-Spectrum Antiviral Activity against Coronaviruses. ACS Infectious Diseases, 7 (3), 586–597. https://doi.org/10.1021/acsinfecdis.0c00761
- Kneller, D. W., Li, H., Phillips, G., Weiss, K. L., Zhang, Q., Arnould, M. A. et al. (2022). Covalent narlaprevir- and boceprevir-derived hybrid inhibitors of SARS-CoV-2 main protease. Nature Communications, 13 (1), 2268. https://doi.org/10.1038/s41467-022-29915-z
- Joyce, R. P., Hu, V. W., Wang, J. (2022). The history, mechanism, and perspectives of nirmatrelvir (PF-07321332): an orally bioavailable main protease inhibitor used in combination with ritonavir to reduce COVID-19-related hospitalizations. Medicinal Chemistry Research, 31 (10), 1637–1646. https://doi.org/10.1007/s00044-022-02951-6
- Tan, H., Hu, Y., Jadhav, P., Tan, B., Wang, J. (2022). Progress and Challenges in Targeting the SARS-CoV-2 Papain-like Protease. Journal of Medicinal Chemistry, 65 (11), 7561–7580. https://doi.org/10.1021/acs.jmedchem.2c00303
- Osipiuk, J., Azizi, S.-A., Dvorkin, S., Endres, M., Jedrzejczak, R., Jones, K. A. et al. (2021). Structure of papain-like protease from SARS-CoV-2 and its complexes with non-covalent inhibitors. Nature Communications, 12 (1), 743. https://doi.org/10.1038/s41467-021-21060-3
- Ton, A.-T., Pandey, M., Smith, J. R., Ban, F., Fernandez, M., Cherkasov, A. (2022). Targeting SARS-CoV-2 papain-like protease in the postvaccine era. Trends in Pharmacological Sciences, 43 (11), 906–919. https://doi.org/10.1016/j.tips.2022.08.008
- Rut, W., Lv, Z., Zmudzinski, M., Patchett, S., Nayak, D., Snipas, S. J. et al. (2020). Activity profiling and crystal structures of inhibitor-bound SARS-CoV-2 papain-like protease: A framework for anti–COVID-19 drug design. Science Advances, 6 (42), eabd4596. https://doi.org/10.1126/sciadv.abd4596
- Gao, X., Qin, B., Chen, P., Zhu, K., Hou, P., Wojdyla, J. A., Wang, M., Cui, S. (2021). Crystal structure of SARS-CoV-2 papain-like protease. Acta Pharmaceutica Sinica B, 11 (1), 237–245. https://doi.org/10.1016/j.apsb.2020.08.014
- Ullrich, S., Nitsche, C. (2022). SARS‐CoV‐2 Papain‐Like Protease: Structure, Function and Inhibition. ChemBioChem, 23 (19). https://doi.org/10.1002/cbic.202200327
- Yuan, S., Gao, X., Tang, K., Cai, J.-P., Hu, M., Luo, P. et al. (2022). Targeting papain-like protease for broad-spectrum coronavirus inhibition. Protein & Cell, 13 (12), 940–953. https://doi.org/10.1007/s13238-022-00909-3
- Wang, Q., Chen, G., He, J., Li, J., Xiong, M., Su, H. et al. (2023). Structure-Based Design of Potent Peptidomimetic Inhibitors Covalently Targeting SARS-CoV-2 Papain-like Protease. International Journal of Molecular Sciences, 24 (10), 8633. https://doi.org/10.3390/ijms24108633
- Yu, W., Zhao, Y., Ye, H., Wu, N., Liao, Y., Chen, N. et al. (2022). Structure-Based Design of a Dual-Targeted Covalent Inhibitor Against Papain-like and Main Proteases of SARS-CoV-2. Journal of Medicinal Chemistry, 65 (24), 16252–16267. https://doi.org/10.1021/acs.jmedchem.2c00954
- Santos, L. H., Kronenberger, T., Almeida, R. G., Silva, E. B., Rocha, R. E. O., Oliveira, J. C. et al. (2022). Structure-Based Identification of Naphthoquinones and Derivatives as Novel Inhibitors of Main Protease Mpro and Papain-like Protease PLpro of SARS-CoV-2. Journal of Chemical Information and Modeling, 62 (24), 6553–6573. https://doi.org/10.1021/acs.jcim.2c00693
- Reddy, A. S., Zhang, S. (2013). Polypharmacology: drug discovery for the future. Expert Review of Clinical Pharmacology, 6 (1), 41–47. https://doi.org/10.1586/ecp.12.74
- Sander, T., Freyss, J., von Korff, M., Rufener, C. (2015). DataWarrior: An Open-Source Program For Chemistry Aware Data Visualization And Analysis. Journal of Chemical Information and Modeling, 55 (2), 460–473. https://doi.org/10.1021/ci500588j
- Wolber, G., Langer, T. (2005). LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters. Journal of Chemical Information and Modeling, 45 (1), 160–169. https://doi.org/10.1021/ci049885e
- Goodsell, D. S., Morris, G. M., Olson, A. J. (1996). Automated docking of flexible ligands: Applications of AutoDock. Journal of Molecular Recognition, 9(1), 1–5. https://doi.org/10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6
- Goodsell, D. S., Sanner, M. F., Olson, A. J., Forli, S. (2020). The AutoDock suite at 30. Protein Science, 30 (1), 31–43. https://doi.org/10.1002/pro.3934
- Kumar, T. D. A. (2022). Drug Design. A Conceptual Overview. Abingdon: CRC Press. https://doi.org/10.1201/9781003298755
- La Monica, G., Bono, A., Lauria, A., Martorana, A. (2022). Targeting SARS-CoV-2 Main Protease for Treatment of COVID-19: Covalent Inhibitors Structure-Activity Relationship Insights and Evolution Perspectives. Journal of Medicinal Chemistry, 65 (19), 12500–12534. https://doi.org/10.1021/acs.jmedchem.2c01005
- Bucherer, H. Th., Lieb, V. A. (1934). Über die Bildung substituierter Hydantoine aus Aldehyden und Ketonen. Synthese von Hydantoinen. Journal Für Praktische Chemie, 141 (1-2), 5–43. https://doi.org/10.1002/prac.19341410102
- Konnert, L., Lamaty, F., Martinez, J., Colacino, E. (2017). Recent Advances in the Synthesis of Hydantoins: The State of the Art of a Valuable Scaffold. Chemical Reviews, 117 (23), 13757–13809. https://doi.org/10.1021/acs.chemrev.7b00067
- Kalník, M., Gabko, P., Bella, M., Koóš, M. (2021). The Bucherer-Bergs Multicomponent Synthesis of Hydantoins – Excellence in Simplicity. Molecules, 26 (13), 4024. https://doi.org/10.3390/molecules26134024
- Park, E. J., Lee, S., Chang, S. (2010). Acetone Cyanohydrin as a Source of HCN in the Cu-Catalyzed Hydrocyanation of α-Aryl Diazoacetates. The Journal of Organic Chemistry, 75 (8), 2760–2762. https://doi.org/10.1021/jo100356d
- Cho, S., Kim, S.-H., Shin, D. (2019). Recent applications of hydantoin and thiohydantoin in medicinal chemistry. European Journal of Medicinal Chemistry, 164, 517–545. https://doi.org/10.1016/j.ejmech.2018.12.066
- Ölmez, N. A., Waseer, F. (2020). New Potential Biologically Active Compounds: Synthesis and Characterization of Urea and Thiourea Derivativpes Bearing 1,2,4-oxadiazole Ring. Current Organic Synthesis, 17 (7), 525–534. https://doi.org/10.2174/1570179417666200417112106
- Zhu, L., Zeng, H., Liu, D., Fu, Y., Wu, Q., Song, B., Gan, X. (2020). Design, synthesis, and biological activity of novel 1,2,4-oxadiazole derivatives. BMC Chemistry, 14 (1), 68. https://doi.org/10.1186/s13065-020-00722-1
- Perekhoda, L., Suleiman, M., Podolsky, I., Semenets, A., Kobzar, N., Yaremenko, V. et al. (2024). Synthesis and nootropic activity prediction of some 4-(aminomethyl)-1-benzylpyrrolidin-2-one derivatives structurally related with nebracetam. ScienceRise: Pharmaceutical Science, 4 (50), 23–34. https://doi.org/10.15587/2519-4852.2024.310731
- Semenets, A. P., Suleiman, M. M., Fedosov, A. I., Shtrygol, S. Y., Havrylov, I. O., Mishchenko, M. V. et al. (2022). Synthesis, docking, and biological evaluation of novel 1-benzyl-4-(4-(R)-5-sulfonylidene-4,5-dihydro-1H-1,2,4-triazol-3-yl)pyrrolidin-2-ones as potential nootropic agents. European Journal of Medicinal Chemistry, 244, 114823. https://doi.org/10.1016/j.ejmech.2022.114823
- Shintani, Y., Kato, K., Kawami, M., Takano, M., Kumamoto, T. (2021). Direct N1-Selective Alkylation of Hydantoins Using Potassium Bases. Chemical and Pharmaceutical Bulletin, 69 (4), 407–410. https://doi.org/10.1248/cpb.c20-00857
##submission.downloads##
Опубліковано
Як цитувати
Номер
Розділ
Ліцензія
Авторське право (c) 2025 Larysa Yevsieieva, Alexander Kyrychenko, Pavlo Trostianko, Volodymyr Ivanov, Sergiy M. Kovalenko, Oleg Kalugin

Ця робота ліцензується відповідно до Creative Commons Attribution 4.0 International License.
Наше видання використовує положення про авторські права Creative Commons CC BY для журналів відкритого доступу.








