Design, synthesis, molecular docking, and antioxidant properties of a series of new S-derivatives of ((1,2,4-triazol-3(2H)-yl)methyl)thiopyrimidines
DOI:
https://doi.org/10.15587/2519-4852.2025.312075Keywords:
1,2,4-triazole derivatives, thiopyrimidines, molecular docking, DPPHantioxidant activityAbstract
The aim of our work is to synthesize new S-derivatives in the series of ((1,2,4-triazol-3(2H)-yl)methyl)thiopyrimidines and to study their antioxidant activity, to identify the most promising compound using molecular docking and kinetic parameters.
Materials and methods. The 1H and 13C NMR spectra were recorded on a Bruker AC-500 spectrometer. LC-MS was recorded on an Agilent 1260 Infinity HPLC system equipped with a diode-array detector and proton ionization. Elemental analysis (C, H, N, S) was performed on an ELEMENTAR vario EL cube. Molecular docking was performed using the AutoDock 4.2.6 program. Free radical absorption was measured using the 1,1-diphenyl-2-picrylhydrazyl (DPPH) free radical assay.
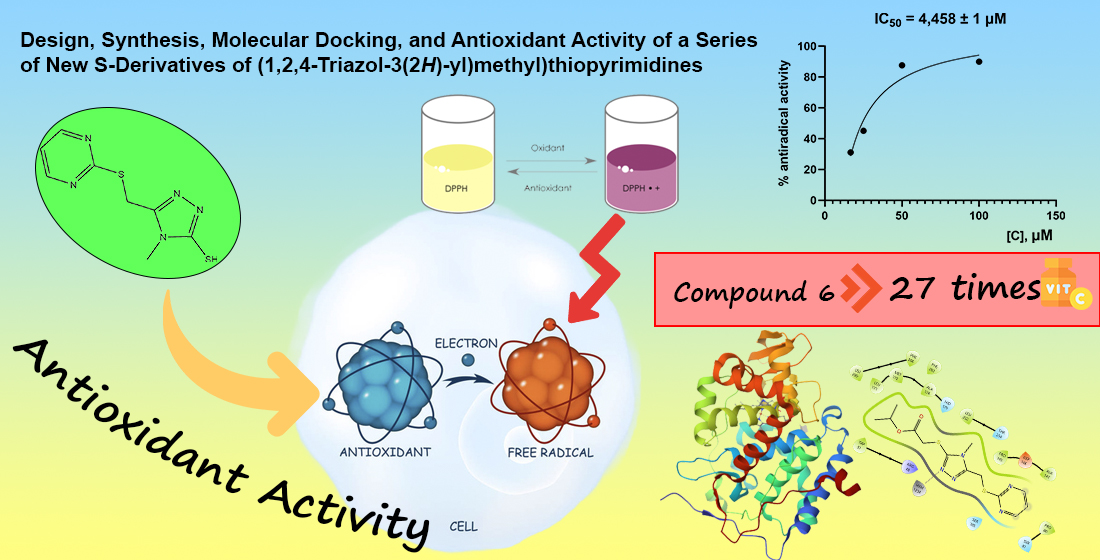
Results. A series of S-acyl derivatives of 4-methyl-5-((pyrimidin-2-ylthio)methyl)-4H-1,2,4-triazole-3-thiols were synthesized through alkylation and subsequent cyclization. The structure of the obtained compounds was confirmed by ¹H and ¹³C NMR spectroscopy. Antiradical activity was evaluated using the DPPH test, with compound (6) exhibiting the highest activity, surpassing ascorbic acid. Molecular docking with cytochrome c peroxidase (PDB: 2X08) confirmed strong binding interactions, highlighting the potential of these derivatives as antioxidants.
Conclusions. Three compounds (1, 6, 8) exhibited higher activity than the reference drug, the natural antioxidant ascorbic acid. This high activity may be associated with the presence of pharmacophore fragments, particularly the pyrimidine skeleton and the sulfur atom linked to the 1,2,4-triazole. The IC50 for the most active compound was calculated as 4.458±1 µM, which is 27 times more effective than ascorbic acid. Molecular docking results showed that compounds 4 and 6 had the lowest binding energies, making them the most effective compounds in terms of antioxidant activity
References
- Singh, S. (2024). Antioxidant nanozymes as next-generation therapeutics to free radical-mediated inflammatory diseases: A comprehensive review. International Journal of Biological Macromolecules, 260, 129374. https://doi.org/10.1016/j.ijbiomac.2024.129374
- Olfat, N., Ashoori, M., Saedisomeolia, A. (2022). Riboflavin is an antioxidant: a review update. British Journal of Nutrition, 128 (10), 1887–1895. https://doi.org/10.1017/s0007114521005031
- Ungor, D., Gombár, G., Juhász, Á., Samu, G. F., Csapó, E. (2023). Promising Bioactivity of Vitamin B1-Au Nanocluster: Structure, Enhanced Antioxidant Behavior, and Serum Protein Interaction. Antioxidants, 12 (4), 874. https://doi.org/10.3390/antiox12040874
- Bhatnagar, A., Pemawat, G. (2023). Functionalized Pyrimidines: Synthetic Approaches and Biological Activities. A Review. Organic Preparations and Procedures International, 56 (1), 1–18. https://doi.org/10.1080/00304948.2023.2225385
- Alamshany, Z. M., Nossier, E. S. (2024). New thiazole derivatives linked to pyridine, fused pyridine, pyrimidine and thiazolopyrimidine scaffolds with potential dual anticancer and antimicrobial activities: Design, synthesis and docking simulation. Journal of Molecular Structure, 1316, 138973. https://doi.org/10.1016/j.molstruc.2024.138973
- Bafail, R. S. M., Samman, W. A. (2024). Anti-parkinsonian, anti-inflammatory, anti-microbial, analgesic, anti-hyperglycemic and anticancer activities of poly-fused ring pyrimidine derivatives. Tropical Journal of Pharmaceutical Research, 23 (1), 67–75. https://doi.org/10.4314/tjpr.v23i1.9
- Myriagkou, M., Papakonstantinou, E., Deligiannidou, G.-E., Patsilinakos, A., Kontogiorgis, C., Pontiki, E. (2023). Novel Pyrimidine Derivatives as Antioxidant and Anticancer Agents: Design, Synthesis and Molecular Modeling Studies. Molecules, 28 (9), 3913. https://doi.org/10.3390/molecules28093913
- El-Naggar, M., Hasan, K., Khanfar, M., Shehadi, I. A., El-Awady, R., El-Dein, A. N. et al. (2024). Synthesis, biological assessment and molecular docking study of new sulfur-linked 1,2,4-triazole and 1,2,3-triazole hybrid derivatives as potential DNA gyrase inhibitors. Zeitschrift Für Naturforschung B, 79 (7), 419–429. https://doi.org/10.1515/znb-2024-0012
- Miedviedieva, K. P., Prytula, R. L., Shmatenko, O. P., Bushuieva, I. V., Parchenko, V. V., Kucherenko, L. I., Vasiuk, S. O. (2024). Express quantitative spectrophotometric determination of 2-(((3-(2-fluorophenyl)- 5-thio-4H-1,2,4-triazol-4-yl)imino)methyl)phenol as an active substance of a medicinal product for the treatment of mycoses. Zaporozhye Medical Journal, 26 (1), 59–65. https://doi.org/10.14739/2310-1210.2024.1.291449
- Karpenko, Y., Kusdemir, G., Parchenko, V., Tüzün, B., Taslimi, P., Karatas, O. F. et al. (2023). A biochemistry‐oriented drug design: synthesis, anticancer activity, enzymes inhibition, molecular docking studies of novel 1,2,4-triazole derivatives. Journal of Biomolecular Structure and Dynamics, 42 (3), 1220–1236. https://doi.org/10.1080/07391102.2023.2253906
- Bitounis, D., Jacquinet, E., Rogers, M. A., Amiji, M. M. (2024). Strategies to reduce the risks of mRNA drug and vaccine toxicity. Nature Reviews Drug Discovery, 23 (4), 281–300. https://doi.org/10.1038/s41573-023-00859-3
- Kaplancıklı, Z., Yurttas, L., Turan-Zitouni, G., Özdemir, A., Göger, G., Demirci, F., Mohsen, U. (2013). Synthesis and Antimicrobial Activity of New Pyrimidine-Hydrazones. Letters in Drug Design & Discovery, 1 (1), 76–81. https://doi.org/10.2174/15701808113109990037
- Pachuta-Stec, A. (2022). Antioxidant Activity of 1,2,4-Triazole and its Derivatives: A Mini-Review. Mini-Reviews in Medicinal Chemistry, 22 (7), 1081–1094. https://doi.org/10.2174/1389557521666210401091802
- Karpenko, Yu. V., Panasenko, O. I., Kulish, S. M., Domnich, A. V. (2023). Synthesis and acute toxicity of new S-derivatives (1,2,4-triazole-3(2H)-yl)methyl) thiopyrimidines. Current Issues in Pharmacy and Medicine: Science and Practice, 16 (2), 158–164. https://doi.org/10.14739/2409-2932.2023.2.274586
- Pham, Q. M., Le, T. T. H., Pham, T. H. M., Tran, Q. T., Do, T. L., Vu, T. T. L., Pham, Q. L. (2022). Molecular docking tutorial using AutoDock 4.2.6 on SARS-CoV-2 main protease for beginner. Vietnam Journal of Science and Technology, 60 (6), 929–947. https://doi.org/10.15625/2525-2518/16459
- Murphy, E. J., Metcalfe, C. L., Basran, J., Moody, P. C. E., Raven, E. L. (2008). Engineering the Substrate Specificity and Reactivity of a Heme Protein: Creation of an Ascorbate Binding Site in Cytochrome c Peroxidase. Biochemistry, 47 (52), 13933–13941. https://doi.org/10.1021/bi801480r
- Aallaei, M., Molaakbari, E., Mostafavi, P., Salarizadeh, N., Maleksah, R. E., & Afzali, D. (2022). Investigation of Cu metal nanoparticles with different morphologies to inhibit SARS-CoV-2 main protease and spike glycoprotein using Molecular Docking and Dynamics Simulation. Journal of Molecular Structure, 1253, 132301. https://doi.org/10.1016/j.molstruc.2021.132301
- Gulcin, İ., Alwasel, S. H. (2023). DPPH Radical Scavenging Assay. Processes, 11 (8), 2248. https://doi.org/10.3390/pr11082248
- Gotsulya, A., Fedotov, S., Zinych, O., Trofimova, T., Brytanova, T. (2023). Synthesis and properties of s-alkyl 4-(4-chlorophenyl)-5-(pyrrole-2-yl)-1,2,4-triazole-3-thiol derivatives. Journal of Faculty of Pharmacy of Ankara University, 47 (3), 1020–1032. https://doi.org/10.33483/jfpau.1280492
- Matuszewska, A., Jaszek, M., Stefaniuk, D., Ciszewski, T., Matuszewski, Ł. (2018). Anticancer, antioxidant, and antibacterial activities of low molecular weight bioactive subfractions isolated from cultures of wood degrading fungus Cerrena unicolor. PLOS ONE, 13 (6), e0197044. https://doi.org/10.1371/journal.pone.0197044
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Yuriy Karpenko, Kateryna Medvedeva, Andrii Solomennyi, Olga Rudenko, Oleksandr Panasenko, Volodymyr Parchenko, Svitlana Vasyuk

This work is licensed under a Creative Commons Attribution 4.0 International License.
Our journal abides by the Creative Commons CC BY copyright rights and permissions for open access journals.







