Implementation of machine learning models to determine the appropriate model for protein function prediction
DOI:
https://doi.org/10.15587/1729-4061.2022.263270Keywords:
protein function prediction, classification, neural networks, ProtCNN, bidirectional long short-term memory (BiLSTM)Abstract
Predicting the function of proteins is a crucial part of genome annotation, which can help in solving a wide range of biological problems. Many methods are available to predict the functions of proteins. However, except for sequence, most features are difficult to obtain or are not available for many proteins, which limits their scope. In addition, the performance of sequence-based feature prediction methods is often lower than that of methods that involve multiple features, and protein feature prediction can be time-consuming. Recent advances in this field are associated with the development of machine learning, which shows great progress in solving the problem of predicting protein functions. Today, however, most protein sequences have the status of «uncharacterized» or «putative».
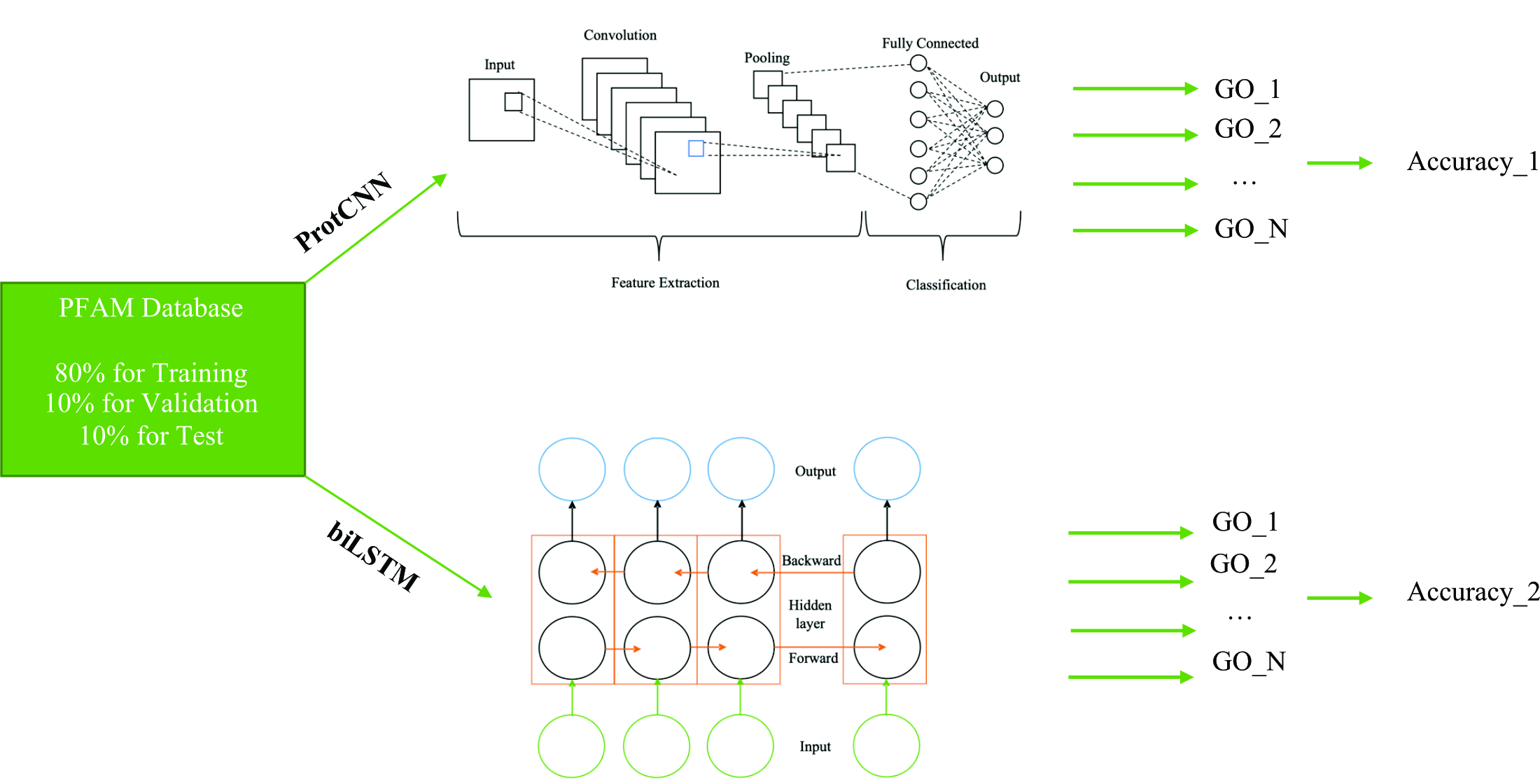
The need to assess the accuracy of identification of protein functions is an urgent task for machine learning approaches used to predict protein functions. In this study, the performance of two popular function prediction algorithms (ProtCNN and BiLSTM) was assessed from two perspectives and the procedures for building these models were described.
As a result of the study of Pfam families, ProtCNN achieves an accuracy rate of 0.988 % and bidirectional LSTM has an accuracy rate of 0.9506 %. The use of the Pfam dataset allowed increasing the classification accuracy due to the large training dataset. The quality of the prediction increases with a large amount of training data.
The study demonstrated that machine learning algorithms can be used as an effective tool for building protein function prediction models, in particular, the CNN network can be adapted as an accurate tool for annotating protein functions in the presence of large datasets.
References
- Gabaldon, T., Huynen, M. A. (2004). Prediction of protein function and pathways in the genome era. Cellular and Molecular Life Sciences (CMLS), 61 (7-8), 930–944. doi: https://doi.org/10.1007/s00018-003-3387-y
- du Plessis, L., Skunca, N., Dessimoz, C. (2011). The what, where, how and why of gene ontology--a primer for bioinformaticians. Briefings in Bioinformatics, 12 (6), 723–735. doi: https://doi.org/10.1093/bib/bbr002
- Barrell, D., Dimmer, E., Huntley, R. P., Binns, D., O’Donovan, C., Apweiler, R. (2009). The GOA database in 2009--an integrated Gene Ontology Annotation resource. Nucleic Acids Research, 37, D396–D403. doi: https://doi.org/10.1093/nar/gkn803
- Piovesan, D., Giollo, M., Leonardi, E., Ferrari, C., Tosatto, S. C. E. (2015). INGA: protein function prediction combining interaction networks, domain assignments and sequence similarity. Nucleic Acids Research, 43 (W1), W134–W140. doi: https://doi.org/10.1093/nar/gkv523
- Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N. et. al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Research, 41 (W1), W29–W33. doi: https://doi.org/10.1093/nar/gkt282
- Stephenson, N., Shane, E., Chase, J., Rowland, J., Ries, D., Justice, N. et. al. (2019). Survey of Machine Learning Techniques in Drug Discovery. Current Drug Metabolism, 20 (3), 185–193. doi: https://doi.org/10.2174/1389200219666180820112457
- Lobley, A. E., Nugent, T., Orengo, C. A., Jones, D. T. (2008). FFPred: an integrated feature-based function prediction server for vertebrate proteomes. Nucleic Acids Research, 36, W297–W302. doi: https://doi.org/10.1093/nar/gkn193
- Cozzetto, D., Minneci, F., Currant, H., Jones, D. T. (2016). FFPred 3: feature-based function prediction for all Gene Ontology domains. Scientific Reports, 6 (1). doi: https://doi.org/10.1038/srep31865
- Jung, J., Yi, G., Sukno, S. A., Thon, M. R. (2010). PoGO: Prediction of Gene Ontology terms for fungal proteins. BMC Bioinformatics, 11 (1). doi: https://doi.org/10.1186/1471-2105-11-215
- Törönen, P., Medlar, A., Holm, L. (2018). PANNZER2: a rapid functional annotation web server. Nucleic Acids Research, 46 (W1), W84–W88. doi: https://doi.org/10.1093/nar/gky350
- You, R., Huang, X., Zhu, S. (2018). DeepText2GO: Improving large-scale protein function prediction with deep semantic text representation. Methods, 145, 82–90. doi: https://doi.org/10.1016/j.ymeth.2018.05.026
- You, R., Yao, S., Xiong, Y., Huang, X., Sun, F., Mamitsuka, H., Zhu, S. (2019). NetGO: improving large-scale protein function prediction with massive network information. Nucleic Acids Research, 47 (W1), W379–W387. doi: https://doi.org/10.1093/nar/gkz388
- Kulmanov, M., Khan, M. A., Hoehndorf, R. (2017). DeepGO: predicting protein functions from sequence and interactions using a deep ontology-aware classifier. Bioinformatics, 34 (4), 660–668. doi: https://doi.org/10.1093/bioinformatics/btx624
- Cai, Y., Wang, J., Deng, L. (2020). SDN2GO: An Integrated Deep Learning Model for Protein Function Prediction. Frontiers in Bioengineering and Biotechnology, 8. doi: https://doi.org/10.3389/fbioe.2020.00391
- Du, Z., He, Y., Li, J., Uversky, V. N. (2020). DeepAdd: Protein function prediction from k-mer embedding and additional features. Computational Biology and Chemistry, 89, 107379. doi: https://doi.org/10.1016/j.compbiolchem.2020.107379
- Zhang, F., Song, H., Zeng, M., Wu, F.-X., Li, Y., Pan, Y., Li, M. (2021). A Deep Learning Framework for Gene Ontology Annotations With Sequence- and Network-Based Information. IEEE/ACM Transactions on Computational Biology and Bioinformatics, 18 (6), 2208–2217. doi: https://doi.org/10.1109/tcbb.2020.2968882
- Spalević, S., Veličković, P., Kovačević, J., Nikolić, M. (2020). Hierarchical Protein Function Prediction with Tail-GNNs. arXiv. doi: https://doi.org/10.48550/arXiv.2007.12804
- LeCun, Y., Bengio, Y., Hinton, G. (2015). Deep learning. Nature, 521 (7553), 436–444. doi: https://doi.org/10.1038/nature14539
- Cao, R., Freitas, C., Chan, L., Sun, M., Jiang, H., Chen, Z. (2017). ProLanGO: Protein Function Prediction Using Neural Machine Translation Based on a Recurrent Neural Network. Molecules, 22 (10), 1732. doi: https://doi.org/10.3390/molecules22101732
- Jiang, Y., Oron, T. R., Clark, W. T., Bankapur, A. R., D’Andrea, D., Lepore, R. et. al. (2016). An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Biology, 17 (1). doi: https://doi.org/10.1186/s13059-016-1037-6
- Pearson, W. R. (2015). Protein Function Prediction: Problems and Pitfalls. Current Protocols in Bioinformatics, 51 (1). doi: https://doi.org/10.1002/0471250953.bi0412s51
- UniProt: the universal protein knowledgebase (2016). Nucleic Acids Research, 45 (D1), D158–D169. doi: https://doi.org/10.1093/nar/gkw1099
- Pfam 35.0 is released. Xfam Blog. Available at: https://xfam.wordpress.com/2021/11/19/pfam-35-0-is-released/
- Bileschi, M. L., Belanger, D., Bryant, D., Sanderson, T., Carter, B., Sculley, D. et. al. (2019). Using Deep Learning to Annotate the Protein Universe. bioRxiv. doi: https://doi.org/10.1101/626507
- Vu, T. T. D., Jung, J. (2021). Protein function prediction with gene ontology: from traditional to deep learning models. PeerJ, 9, e12019. doi: https://doi.org/10.7717/peerj.12019
- Abduljabbar, R. L., Dia, H., Tsai, P.-W. (2021). Unidirectional and Bidirectional LSTM Models for Short-Term Traffic Prediction. Journal of Advanced Transportation, 2021, 1–16. doi: https://doi.org/10.1155/2021/5589075
- Kurtukova, A. V., Romanov, A. S. (2019). Modeling the neural network architecture to identify the author of the source code. Proceedings of Tomsk State University of Control Systems and Radioelectronics, 22 (3), 37–42. doi: https://doi.org/10.21293/1818-0442-2019-22-3-37-42
- Deen, A., Gayanchandani, M. (2019). Protein Function Prediction using SVM Kernel Approach. International Journal of Scientific & Engineering Research, 10 (7), 1995–2000. Available at: https://www.ijser.org/researchpaper/Protein-Function-Prediction-using-SVM-Kernel-Approach.pdf
- Kingma, D. P., Ba, J. (2014). Adam: A Method for Stochastic Optimization. 3rd International Conference for Learning Representations. San Diego. doi: https://doi.org/10.48550/arXiv.1412.6980
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2022 Yekaterina Golenko, Aisulu Ismailova, Anargul Shaushenova, Zhazira Mutalova, Damir Dossalyanov, Aliya Ainagulova, Akgul Naizagarayeva

This work is licensed under a Creative Commons Attribution 4.0 International License.
The consolidation and conditions for the transfer of copyright (identification of authorship) is carried out in the License Agreement. In particular, the authors reserve the right to the authorship of their manuscript and transfer the first publication of this work to the journal under the terms of the Creative Commons CC BY license. At the same time, they have the right to conclude on their own additional agreements concerning the non-exclusive distribution of the work in the form in which it was published by this journal, but provided that the link to the first publication of the article in this journal is preserved.
A license agreement is a document in which the author warrants that he/she owns all copyright for the work (manuscript, article, etc.).
The authors, signing the License Agreement with TECHNOLOGY CENTER PC, have all rights to the further use of their work, provided that they link to our edition in which the work was published.
According to the terms of the License Agreement, the Publisher TECHNOLOGY CENTER PC does not take away your copyrights and receives permission from the authors to use and dissemination of the publication through the world's scientific resources (own electronic resources, scientometric databases, repositories, libraries, etc.).
In the absence of a signed License Agreement or in the absence of this agreement of identifiers allowing to identify the identity of the author, the editors have no right to work with the manuscript.
It is important to remember that there is another type of agreement between authors and publishers – when copyright is transferred from the authors to the publisher. In this case, the authors lose ownership of their work and may not use it in any way.








