Development of data-efficient training techniques for detection and segmentation models in atrial septum defect analysis
DOI:
https://doi.org/10.15587/1729-4061.2024.312621Keywords:
deep learning, SegFormer, echocardiography, image segmentation, YOLOv7, data augmentationAbstract
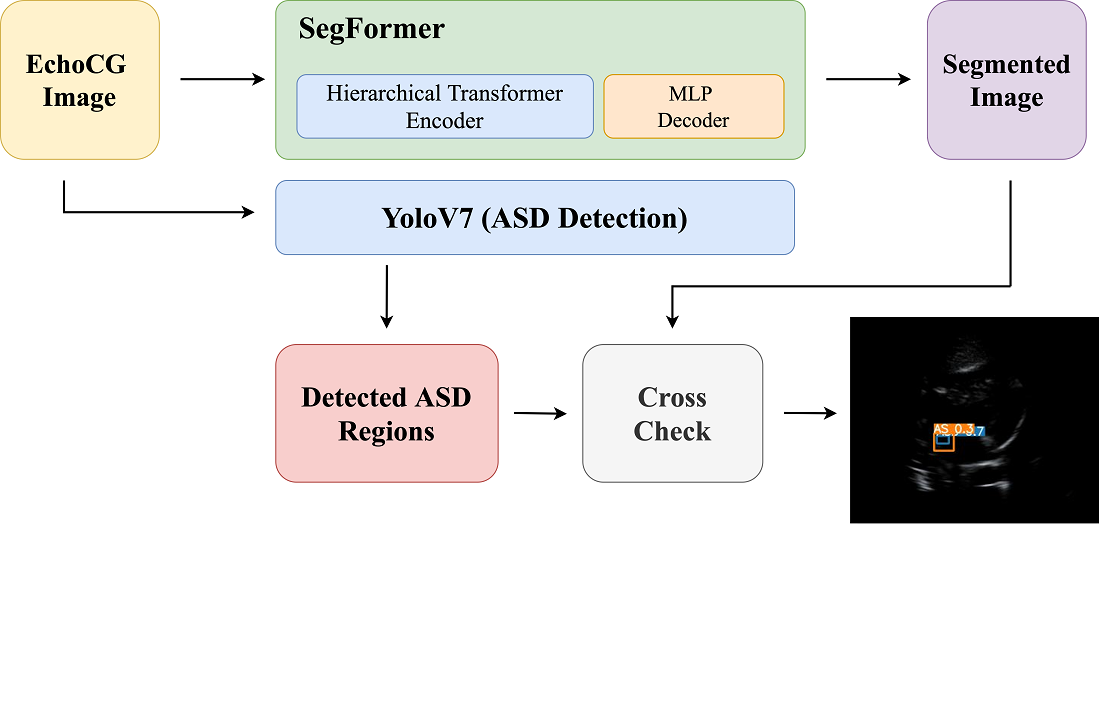
The object of this research is to develop a data-efficient pipeline for the detection of atrial septal defects (ASDs) using echocardiographic images. ASDs are common congenital heart defects that can lead to serious health issues if not diagnosed early. Rising mortality rates due to undetected ASDs highlight the urgent need for improved diagnostic methods. To address the problem of limited annotated medical data hindering accurate detection models, this study fine-tuned the SegFormer model for precise segmentation of cardiac structures in echocardiography images, focusing on the four-chamber heart view essential for ASD detection. By integrating SegFormer with the YOLOv7 detection model, known for real-time object detection, the ASD regions within the segmented heart structures were accurately identified. This cross-referencing ensures anatomically accurate diagnoses and reduces false positives. The study results demonstrate that despite limited data, the integrated method achieves high accuracy and speed, outperforming traditional models. This improvement is explained by the synergy between SegFormer’s transformer-based segmentation and YOLOv7’s efficient detection capabilities. The distinctive feature of our approach is the successful integration of these models in a data-efficient manner, enabling effective ASD detection even with scarce data. The scope of practical use includes deployment in clinical settings with limited resources, requiring only echocardiographic equipment and basic computational resources. By providing clinicians with a reliable tool for ASD detection, the study supports timely interventions in pediatric cardiology, ultimately improving patient outcomes and enhancing care consistency
References
- Williams, M. R., Perry, J. C. (2018). Arrhythmias and conduction disorders associated with atrial septal defects. Journal of Thoracic Disease, 10 (S24), S2940–S2944. https://doi.org/10.21037/jtd.2018.08.27
- Liu, Y., Huang, Q., Han, X., Liang, T., Zhang, Z., Lu, X. et al. (2024). Atrial Septal Defect Detection in Children Based on Ultrasound Video Using Multiple Instances Learning. Journal of Imaging Informatics in Medicine, 37 (3), 965–975. https://doi.org/10.1007/s10278-024-00987-1
- Mertens, L., Friedberg, M. K. (2009). The gold standard for noninvasive imaging in congenital heart disease: echocardiography. Current Opinion in Cardiology, 24 (2), 119–124. https://doi.org/10.1097/hco.0b013e328323d86f
- Sadeghpour, A., Alizadehasl, A. (2018). Echocardiography in the Critical Care Unit. Case-Based Textbook of Echocardiography, 423–430. https://doi.org/10.1007/978-3-319-67691-3_32
- Lancellotti, P., Price, S., Edvardsen, T., Cosyns, B., Neskovic, A. N., Dulgheru, R. et al. (2014). The use of echocardiography in acute cardiovascular care: Recommendations of the European Association of Cardiovascular Imaging and the Acute Cardiovascular Care Association. European Heart Journal - Cardiovascular Imaging, 16 (2), 119–146. https://doi.org/10.1093/ehjci/jeu210
- Tiver, K. D., Horsfall, M., Swan, A., De Pasquale, C., Horsfall, E., Chew, D. P., De Pasquale, C. G. (2022). Accuracy of Highly Limited Echocardiographic Screening Images for Determining a Structurally Normal Heart: The Quick-Six Study. Heart, Lung and Circulation, 31 (4), 462–468. https://doi.org/10.1016/j.hlc.2021.08.021
- Sicari, R., Gargani, L., Wiecek, A., Covic, A., Goldsmith, D., Suleymanlar, G. et al. (2012). The use of echocardiography in observational clinical trials: the EURECA-m registry. Nephrology Dialysis Transplantation, 28 (1), 19–23. https://doi.org/10.1093/ndt/gfs399
- Grenon, V., Szymonifka, J., Adler-Milstein, J., Ross, J., Sarkar, U. (2023). Factors Associated With Diagnostic Error: An Analysis of Closed Medical Malpractice Claims. Journal of Patient Safety, 19 (3), 211–215. https://doi.org/10.1097/pts.0000000000001105
- Li, Y., Liu, Z., Lai, Q., Li, S., Guo, Y., Wang, Y. et al. (2022). ESA-UNet for assisted diagnosis of cardiac magnetic resonance image based on the semantic segmentation of the heart. Frontiers in Cardiovascular Medicine, 9. https://doi.org/10.3389/fcvm.2022.1012450
- Madani, A., Arnaout, R., Mofrad, M., Arnaout, R. (2018). Fast and accurate view classification of echocardiograms using deep learning. Npj Digital Medicine, 1 (1). https://doi.org/10.1038/s41746-017-0013-1
- Shamir, O. (2018). Are resnets provably better than linear predictors. arXiv. https://doi.org/10.48550/arXiv.1804.06739
- Li, Y.-Z., Wang, Y., Huang, Y.-H., Xiang, P., Liu, W.-X., Lai, Q.-Q. et al. (2023). RSU-Net: U-net based on residual and self-attention mechanism in the segmentation of cardiac magnetic resonance images. Computer Methods and Programs in Biomedicine, 231, 107437. https://doi.org/10.1016/j.cmpb.2023.107437
- Ghorbani, A., Ouyang, D., Abid, A., He, B., Chen, J. H., Harrington, R. A. et al. (2020). Deep learning interpretation of echocardiograms. Npj Digital Medicine, 3 (1). https://doi.org/10.1038/s41746-019-0216-8
- Lim, G. B. (2020). Estimating ejection fraction by video-based AI. Nature Reviews Cardiology, 17 (6), 320–320. https://doi.org/10.1038/s41569-020-0375-y
- Guo, Z., Zhang, Y., Qiu, Z., Dong, S., He, S., Gao, H. et al. (2023). An improved contrastive learning network for semi-supervised multi-structure segmentation in echocardiography. Frontiers in Cardiovascular Medicine, 10. https://doi.org/10.3389/fcvm.2023.1266260
- Vakanski, A., Xian, M. (2021). Evaluation of Complexity Measures for Deep Learning Generalization in Medical Image Analysis. 2021 IEEE 31st International Workshop on Machine Learning for Signal Processing (MLSP), abs 2012 4115, 1–6. https://doi.org/10.1109/mlsp52302.2021.9596501
- Moradi, S., Oghli, M. G., Alizadehasl, A., Shiri, I., Oveisi, N., Oveisi, M. et al. (2019). MFP-Unet: A novel deep learning based approach for left ventricle segmentation in echocardiography. Physica Medica, 67, 58–69. https://doi.org/10.1016/j.ejmp.2019.10.001
- Xie, E., Wang, W., Yu, Z., Anandkumar, A., Alvarez, J. M., Luo, P. (2021). Segformer: Simple and efficient design for semantic segmentation with transformers. arXiv. https://doi.org/10.48550/arXiv.2105.15203
- Liu, X., Yang, X. (2008). Automatic acquisition of the four-chamber view for 3D echocardiography. IEICE Electronics Express, 5 (9), 316–320. https://doi.org/10.1587/elex.5.316
- Ukibassov, B. M., Rakhmetulayeva, S. B., Zhanabekov, Zh. O., Bolshibayeva, A. K., Yasar, A.-U.-H. (2024). Implementation of Anatomy Constrained Contrastive Learning for Heart Chamber Segmentation. Procedia Computer Science, 238, 536–543. https://doi.org/10.1016/j.procs.2024.06.057
- Zhou, Q., Sun, Z., Wang, L., Kang, B., Zhang, S., Wu, X. (2023). Mixture lightweight transformer for scene understanding. Computers and Electrical Engineering, 108, 108698. https://doi.org/10.1016/j.compeleceng.2023.108698
- Silvestry, F. E., Cohen, M. S., Armsby, L. B., Burkule, N. J., Fleishman, C. E., Hijazi, Z. M., Lang, R. M. et al. (2015). Guidelines for the Echocardiographic Assessment of Atrial Septal Defect and Patent Foramen Ovale: From the American Society of Echocardiography and Society for Cardiac Angiography and Interventions. Journal of the American Society of Echocardiography, 28 (8), 910–958. https://doi.org/10.1016/j.echo.2015.05.015
- Rakhmetulayeva, S. B., Bolshibayeva, A. K., Mukasheva, A. K., Ukibassov, B. M., Zhanabekov, Zh. O., Diaz, D. (2023). Machine learning methods and algorithms for predicting congenital heart pathologies. 2023 IEEE 17th International Conference on Application of Information and Communication Technologies (AICT). https://doi.org/10.1109/aict59525.2023.10313184
- Jwaid, W. M., Al-Husseini, Z. S. M., Sabry, A. H. (2021). Development of brain tumor segmentation of magnetic resonance imaging (MRI) using U-Net deep learning. Eastern-European Journal of Enterprise Technologies, 4 (9 (112)), 23–31. https://doi.org/10.15587/1729-4061.2021.238957
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Sabina Rakhmetulayeva, Baubek Ukibassov, Zhandos Zhanabekov, Aigerim Bolshibayeva

This work is licensed under a Creative Commons Attribution 4.0 International License.
The consolidation and conditions for the transfer of copyright (identification of authorship) is carried out in the License Agreement. In particular, the authors reserve the right to the authorship of their manuscript and transfer the first publication of this work to the journal under the terms of the Creative Commons CC BY license. At the same time, they have the right to conclude on their own additional agreements concerning the non-exclusive distribution of the work in the form in which it was published by this journal, but provided that the link to the first publication of the article in this journal is preserved.
A license agreement is a document in which the author warrants that he/she owns all copyright for the work (manuscript, article, etc.).
The authors, signing the License Agreement with TECHNOLOGY CENTER PC, have all rights to the further use of their work, provided that they link to our edition in which the work was published.
According to the terms of the License Agreement, the Publisher TECHNOLOGY CENTER PC does not take away your copyrights and receives permission from the authors to use and dissemination of the publication through the world's scientific resources (own electronic resources, scientometric databases, repositories, libraries, etc.).
In the absence of a signed License Agreement or in the absence of this agreement of identifiers allowing to identify the identity of the author, the editors have no right to work with the manuscript.
It is important to remember that there is another type of agreement between authors and publishers – when copyright is transferred from the authors to the publisher. In this case, the authors lose ownership of their work and may not use it in any way.








