Automated extraction of key parameters and detection of inconsistencies in clinical documentation using large language models
DOI:
https://doi.org/10.15587/1729-4061.2025.337915Keywords:
clinical trials, large language models, clinical documentation, data miningAbstract
This study investigates unstructured text data on clinical trials. The task addressed relates to the fact that analyzing such data involves a laborious and error-prone process, hard-to-tackle even for specialists. In turn, this leads to an increase in the duration of studies and delays in the release of new drugs to the market.
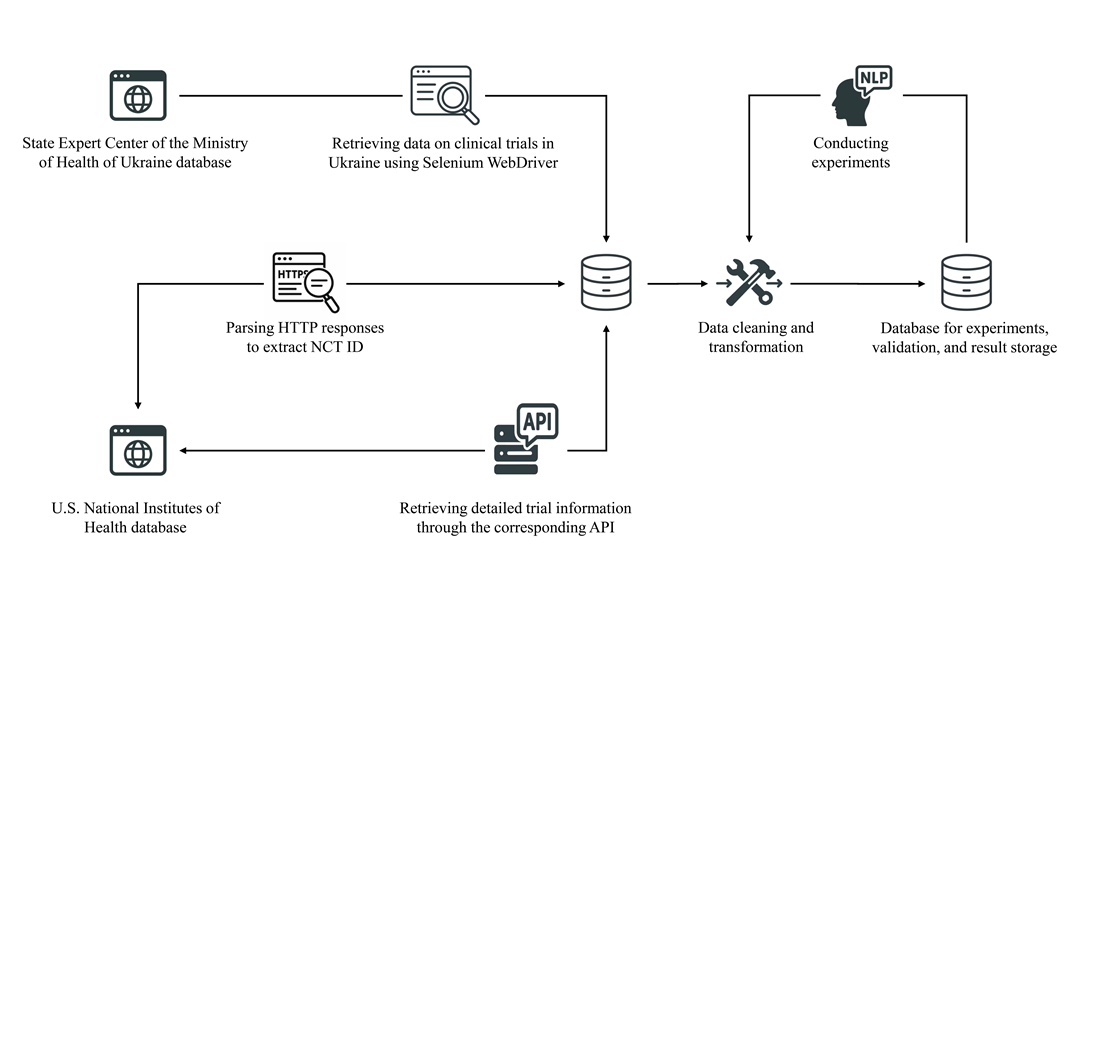
This work reports an approach to constructing a dataset on clinical trials, as well as subsequent extraction of key information using state-of-the-art large language models. A study was conducted on extracting such indicators as the eligible gender of participants, a research phase, as well as the study's therapeutic area. A total of 11,703 experiments were performed, most of which achieved high results. In particular, the average values when using the GPT-4o-mini model were as follows: F1-measure – 0.92; accuracy – 0.98; recall – 0.99; precision – 0.87.
Extraction of information from clinical documentation in Ukrainian demonstrated similar results compared to English-language counterparts. In some cases, a significant number of false positives were observed, and the indicators were significantly lower (the lowest recorded values: F1-measure – 0.52; accuracy – 0.82; recall – 0.78; precision – 0.35). For such cases, the reasons were analyzed, and the corresponding conclusions and recommendations were formulated.
In addition, the results of the experiments helped identify a number of discrepancies and errors in official registries, which is a vivid example of practical application. Other examples of using the result are the possibility of scaling the technology to additional data types, as well as supporting digital transformation in the medical field. Such results are prerequisites for automating the clinical trial process and accelerating the release of new drugs to the market.
References
- Cascini, F., Beccia, F., Causio, F. A., Melnyk, A., Zaino, A., Ricciardi, W. (2022). Scoping review of the current landscape of AI-based applications in clinical trials. Frontiers in Public Health, 10. https://doi.org/10.3389/fpubh.2022.949377
- Kreimeyer, K., Foster, M., Pandey, A., Arya, N., Halford, G., Jones, S. F. et al. (2017). Natural language processing systems for capturing and standardizing unstructured clinical information: A systematic review. Journal of Biomedical Informatics, 73, 14–29. https://doi.org/10.1016/j.jbi.2017.07.012
- Alexander, M., Solomon, B., Ball, D. L., Sheerin, M., Dankwa-Mullan, I., Preininger, A. M. et al. (2020). Evaluation of an artificial intelligence clinical trial matching system in Australian lung cancer patients. JAMIA Open, 3 (2), 209–215. https://doi.org/10.1093/jamiaopen/ooaa002
- Liu, X., Hersch, G. L., Khalil, I., Devarakonda, M. (2021). Clinical Trial Information Extraction with BERT. 2021 IEEE 9th International Conference on Healthcare Informatics (ICHI). IEEE, 505–506. https://doi.org/10.1109/ichi52183.2021.00092
- Li, J., Wei, Q., Ghiasvand, O., Chen, M., Lobanov, V., Weng, C., Xu, H. (2022). A comparative study of pre-trained language models for named entity recognition in clinical trial eligibility criteria from multiple corpora. BMC Medical Informatics and Decision Making, 22 (S3). https://doi.org/10.1186/s12911-022-01967-7
- Nori, H., Lee, Y. T., Zhang, S., Carignan, D., Edgar, R., Fusi, N. et al. (2023). Can generalist foundation models outcompete special-purpose tuning? Case study in medicine. arXiv preprint. arXiv:2311.16452. https://doi.org/10.48550/arXiv.2311.16452
- Lee, J.-M. (2024). Strategies for integrating ChatGPT and generative AI into clinical studies. Blood Research, 59 (1). https://doi.org/10.1007/s44313-024-00045-3
- Lee, K., Paek, H., Huang, L.-C., Hilton, C. B., Datta, S., Higashi, J. et al. (2024). SEETrials: Leveraging large language models for safety and efficacy extraction in oncology clinical trials. Informatics in Medicine Unlocked, 50, 101589. https://doi.org/10.1016/j.imu.2024.101589
- Wong, A., Plasek, J. M., Montecalvo, S. P., Zhou, L. (2018). Natural Language Processing and Its Implications for the Future of Medication Safety: A Narrative Review of Recent Advances and Challenges. Pharmacotherapy: The Journal of Human Pharmacology and Drug Therapy, 38 (8), 822–841. https://doi.org/10.1002/phar.2151
- Dyyak, I., Horlatch, V., Pasichnyk, T., Pasichnyk, V. (2024). Assessing generative pre-trained transformer 4 in clinical trial inclusion criteria matching. Proceedings of the CEUR Workshop, 3702, 305–316. Available at: https://ceur-ws.org/Vol-3702/paper25.pdf
- Kang, T., Zhang, S., Tang, Y., Hruby, G. W., Rusanov, A., Elhadad, N., Weng, C. (2017). EliIE: An open-source information extraction system for clinical trial eligibility criteria. Journal of the American Medical Informatics Association, 24 (6), 1062–1071. https://doi.org/10.1093/jamia/ocx019
- Zeng, K., Xu, Y., Lin, G., Liang, L., Hao, T. (2021). Automated classification of clinical trial eligibility criteria text based on ensemble learning and metric learning. BMC Medical Informatics and Decision Making, 21 (S2). https://doi.org/10.1186/s12911-021-01492-z
- Kury, F., Butler, A., Yuan, C., Fu, L., Sun, Y., Liu, H. et al. (2020). Chia, a large annotated corpus of clinical trial eligibility criteria. Scientific Data, 7 (1). https://doi.org/10.1038/s41597-020-00620-0
- Thirunavukarasu, A. J., Ting, D. S. J., Elangovan, K., Gutierrez, L., Tan, T. F., Ting, D. S. W. (2023). Large language models in medicine. Nature Medicine, 29 (8), 1930–1940. https://doi.org/10.1038/s41591-023-02448-8
- Veen, D. V., Uden, C. V., Blankemeier, L., Delbrouck, J.-B., Aali, A., Bluethgen, C. et al. (2023). Clinical Text Summarization: Adapting Large Language Models Can Outperform Human Experts. https://doi.org/10.21203/rs.3.rs-3483777/v1
- Luo, X., Chen, F., Zhu, D., Wang, L., Wang, Z., Liu, H. et al. (2024). Potential Roles of Large Language Models in the Production of Systematic Reviews and Meta-Analyses. Journal of Medical Internet Research, 26, e56780. https://doi.org/10.2196/56780
- Datta, S., Lee, K., Paek, H., Manion, F. J., Ofoegbu, N., Du, J. et al. (2023). AutoCriteria: a generalizable clinical trial eligibility criteria extraction system powered by large language models. Journal of the American Medical Informatics Association, 31 (2), 375–385. https://doi.org/10.1093/jamia/ocad218
- Kim, J., Quintana, Y. (2022). Review of the Performance Metrics for Natural Language Systems for Clinical Trials Matching. MEDINFO 2021: One World, One Health – Global Partnership for Digital Innovation. IOS Press Ebooks, 641–644. https://doi.org/10.3233/shti220156
- Jin, Q., Wang, Z., Floudas, C. S., Chen, F., Gong, C., Bracken-Clarke, D. et al. (2024). Matching patients to clinical trials with large language models. Nature Communications, 15 (1). https://doi.org/10.1038/s41467-024-53081-z
- Pasichnyk, V. (2025). Datasets of clinical trials in Ukraine. Figshare. Collection. https://doi.org/10.6084/m9.figshare.c.7887785.v1
- Pasichnyk, V. (2025). Results of key data extraction from clinical trial documentation. Figshare. Dataset. https://doi.org/10.6084/m9.figshare.29378450
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Vasyl Pasichnyk, Vitaliy Horlatch

This work is licensed under a Creative Commons Attribution 4.0 International License.
The consolidation and conditions for the transfer of copyright (identification of authorship) is carried out in the License Agreement. In particular, the authors reserve the right to the authorship of their manuscript and transfer the first publication of this work to the journal under the terms of the Creative Commons CC BY license. At the same time, they have the right to conclude on their own additional agreements concerning the non-exclusive distribution of the work in the form in which it was published by this journal, but provided that the link to the first publication of the article in this journal is preserved.
A license agreement is a document in which the author warrants that he/she owns all copyright for the work (manuscript, article, etc.).
The authors, signing the License Agreement with TECHNOLOGY CENTER PC, have all rights to the further use of their work, provided that they link to our edition in which the work was published.
According to the terms of the License Agreement, the Publisher TECHNOLOGY CENTER PC does not take away your copyrights and receives permission from the authors to use and dissemination of the publication through the world's scientific resources (own electronic resources, scientometric databases, repositories, libraries, etc.).
In the absence of a signed License Agreement or in the absence of this agreement of identifiers allowing to identify the identity of the author, the editors have no right to work with the manuscript.
It is important to remember that there is another type of agreement between authors and publishers – when copyright is transferred from the authors to the publisher. In this case, the authors lose ownership of their work and may not use it in any way.








