Advancing real-time echocardiographic diagnosis with a hybrid deep learning model
DOI:
https://doi.org/10.15587/1729-4061.2024.314845Keywords:
deep learning, machine learning, CNN, YOLOv7, SegFormer, transformer-based modelsAbstract
This research focuses on developing a novel hybrid deep learning architecture designed for real-time analysis of ultrasound heart images. The object of the study is the diagnostic accuracy and efficiency in detecting heart pathologies such as atrial septal defect (ASD) and aortic stenosis (AS) from ultrasound data.
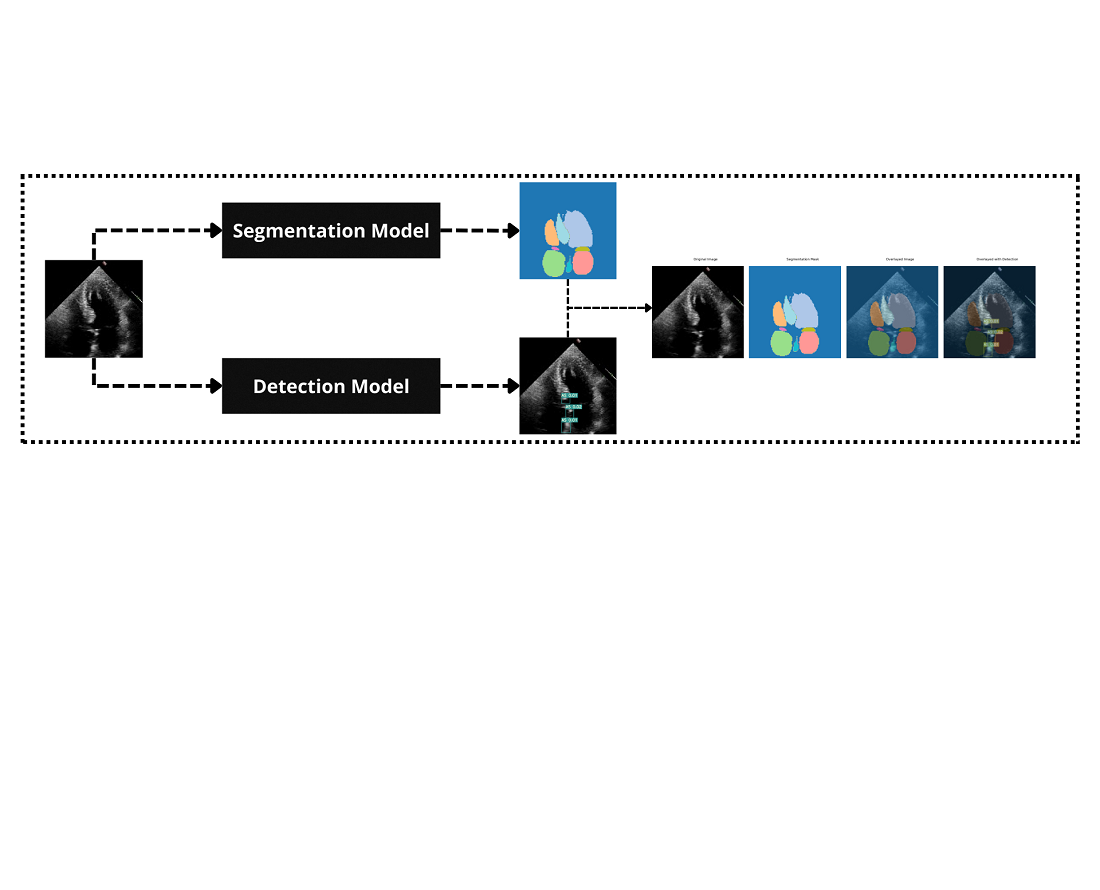
The problem is the insufficient accuracy and generalizability of existing models in real-time cardiac image analysis, which limits their practical clinical application. To solve this, the convolutional neural networks (CNNs), combining local feature extraction was integrated with global contextual understanding of cardiac structures. Additionally, a YOLOv7 for precise segmentation and detection was utilized.
The results demonstrate that the hybrid model achieves an overall diagnostic accuracy of 92 % for ASD detection and 90 % for AS detection, representing a 7 % improvement over the standard YOLOv7 model. These improvements are attributed to the hybrid architecture's ability to simultaneously capture fine-grained anatomical details and broader structural relationships, enhancing the detection of subtle cardiac anomalies.
The findings suggest that combination of CNNs enhances pattern recognition and contextual analysis, leading to better detection of cardiac anomalies. The key features contributing to solving the problem include the hybrid architecture's ability to capture detailed local features and broader structural context simultaneously.
In practical terms, the model can be applied in clinical settings that require real-time cardiac assessment using standard medical imaging equipment. Its computational efficiency and high accuracy make it suitable even in resource-constrained environments, reducing analysis time for clinicians, supporting personalized treatment plans, and potentially improving patient outcomes in cardiology
References
- Cardiovascular diseases (CVDs) (2021). World Health Organization. Available at: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds)
- Nagueh, S. F., Smiseth, O. A., Appleton, C. P., Byrd, B. F., Dokainish, H., Edvardsen, T. et al. (2016). Recommendations for the Evaluation of Left Ventricular Diastolic Function by Echocardiography: An Update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. European Heart Journal – Cardiovascular Imaging, 17 (12), 1321–1360. https://doi.org/10.1093/ehjci/jew082
- Zhou, J., Du, M., Chang, S., Chen, Z. (2021). Artificial intelligence in echocardiography: detection, functional evaluation, and disease diagnosis. Cardiovascular Ultrasound, 19 (1). https://doi.org/10.1186/s12947-021-00261-2
- Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M. et al. (2017). A survey on deep learning in medical image analysis. Medical Image Analysis, 42, 60–88. https://doi.org/10.1016/j.media.2017.07.005
- Shen, D., Wu, G., Suk, H.-I. (2017). Deep Learning in Medical Image Analysis. Annual Review of Biomedical Engineering, 19 (1), 221–248. https://doi.org/10.1146/annurev-bioeng-071516-044442
- Wang, C.-Y., Bochkovskiy, A., Liao, H.-Y. M. (2023). YOLOv7: Trainable Bag-of-Freebies Sets New State-of-the-Art for Real-Time Object Detectors. 2023 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 7464–7475. https://doi.org/10.1109/cvpr52729.2023.00721
- Ho, J., Jain, A., Abbeel, P. (2020). Denoising diffusion probabilistic models. Advances in Neural Information Processing Systems, 33, 6840–6851.
- Zhang, J., Gajjala, S., Agrawal, P., Tison, G. H., Hallock, L. A., Beussink-Nelson, L. et al. (2018). Fully Automated Echocardiogram Interpretation in Clinical Practice. Circulation, 138 (16), 1623–1635. https://doi.org/10.1161/circulationaha.118.034338
- Ouyang, D., He, B., Ghorbani, A., Yuan, N., Ebinger, J., Langlotz, C. P. et al. (2020). Video-based AI for beat-to-beat assessment of cardiac function. Nature, 580 (7802), 252–256. https://doi.org/10.1038/s41586-020-2145-8
- Leclerc, S., Smistad, E., Pedrosa, J., Ostvik, A., Cervenansky, F., Espinosa, F. et al. (2019). Deep Learning for Segmentation Using an Open Large-Scale Dataset in 2D Echocardiography. IEEE Transactions on Medical Imaging, 38 (9), 2198–2210. https://doi.org/10.1109/tmi.2019.2900516
- Lang, R. M., Badano, L. P., Mor-Avi, V., Afilalo, J., Armstrong, A., Ernande, L. et al. (2015). Recommendations for Cardiac Chamber Quantification by Echocardiography in Adults: An Update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. Journal of the American Society of Echocardiography, 28 (1), 1-39.e14. https://doi.org/10.1016/j.echo.2014.10.003
- Knackstedt, C., Bekkers, S. C. A. M., Schummers, G., Schreckenberg, M., Muraru, D., Badano, L. P. et al. (2015). Fully Automated Versus Standard Tracking of Left Ventricular Ejection Fraction and Longitudinal Strain. Journal of the American College of Cardiology, 66 (13), 1456–1466. https://doi.org/10.1016/j.jacc.2015.07.052
- Madani, A., Arnaout, R., Mofrad, M., Arnaout, R. (2018). Fast and accurate view classification of echocardiograms using deep learning. Npj Digital Medicine, 1 (1). https://doi.org/10.1038/s41746-017-0013-1
- Ronneberger, O., Fischer, P., Brox, T. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015, 234–241. https://doi.org/10.1007/978-3-319-24574-4_28
- Oktay, O., Schlemper, J., Folgoc, L. L., Lee, M., Heinrich, M., Misawa, K. et al. (2018). Attention U-Net: Learning where to look for the pancreas. arXiv. https://doi.org/10.48550/arXiv.1804.03999
- Wang, K., Wang, S., Xiong, M., Wang, C., Wang, H. (2021). Non-invasive Assessment of Hepatic Venous Pressure Gradient (HVPG) Based on MR Flow Imaging and Computational Fluid Dynamics. Medical Image Computing and Computer Assisted Intervention – MICCAI 2021, 33–42. https://doi.org/10.1007/978-3-030-87234-2_4
- Mudeng, V., Nisa, W., Sukmananda Suprapto, S. (2022). Computational image reconstruction for multi-frequency diffuse optical tomography. Journal of King Saud University - Computer and Information Sciences, 34 (6), 3527–3538. https://doi.org/10.1016/j.jksuci.2020.12.015
- Nurmaini, S., Sapitri, A. I., Tutuko, B., Rachmatullah, M. N., Rini, D. P., Darmawahyuni, A. et al. (2023). Automatic echocardiographic anomalies interpretation using a stacked residual-dense network model. BMC Bioinformatics, 24 (1). https://doi.org/10.1186/s12859-023-05493-9
- Iriani Sapitri, A., Nurmaini, S., Naufal Rachmatullah, M., Tutuko, B., Darmawahyuni, A., Firdaus, F. et al. (2023). Deep learning-based real time detection for cardiac objects with fetal ultrasound video. Informatics in Medicine Unlocked, 36, 101150. https://doi.org/10.1016/j.imu.2022.101150
- Sutarno, S., Nurmaini, S., Partan, R. U., Sapitri, A. I., Tutuko, B., Naufal Rachmatullah, M. et al. (2022). FetalNet: Low-light fetal echocardiography enhancement and dense convolutional network classifier for improving heart defect prediction. Informatics in Medicine Unlocked, 35, 101136. https://doi.org/10.1016/j.imu.2022.101136
- Saxena, A., Singh, S. P., Gaidhane, V. H. (2022). A deep learning approach for the detection of COVID-19 from chest X-ray images using convolutional neural networks. Advances in Machine Learning & Artificial Intelligence, 3 (2), 52–65. https://doi.org/10.33140/amlai.03.02.01
- Zhou, Q., Sun, Z., Wang, L., Kang, B., Zhang, S., Wu, X. (2023). Mixture lightweight transformer for scene understanding. Computers and Electrical Engineering, 108, 108698. https://doi.org/10.1016/j.compeleceng.2023.108698
- Xie, E., Wang, W., Yu, Z., Anandkumar, A., Alvarez, J. M., Luo, P. (2021). SegFormer: Simple and efficient design for semantic segmentation with transformers. arXiv. https://doi.org/10.48550/arXiv.2105.15203
- Rakhmetulayeva, S. B., Bolshibayeva, A. K., Mukasheva, A. K., Ukibassov, B. M., Zhanabekov, Zh. O., Diaz, D. (2023). Machine learning methods and algorithms for predicting congenital heart pathologies. 2023 IEEE 17th International Conference on Application of Information and Communication Technologies (AICT). https://doi.org/10.1109/aict59525.2023.10313184
- Ukibassov, B. M., Rakhmetulayeva, S. B., Zhanabekov, Zh. O., Bolshibayeva, A. K., Yasar, A.-U.-H. (2024). Implementation of Anatomy Constrained Contrastive Learning for Heart Chamber Segmentation. Procedia Computer Science, 238, 536–543. https://doi.org/10.1016/j.procs.2024.06.057
- Rakhmetulayeva, S., Syrymbet, Z. (2022). Implementation of convolutional neural network for predicting glaucoma from fundus images. Eastern-European Journal of Enterprise Technologies, 6 (2 (120)), 70–77. https://doi.org/10.15587/1729-4061.2022.269229
- He, K., Zhang, X., Ren, S., Sun, J. (2016). Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR). https://doi.org/10.1109/cvpr.2016.90
- Liu, Z., Lin, Y., Cao, Y., Hu, H., Wei, Y., Zhang, Z. et al. (2021). Swin Transformer: Hierarchical Vision Transformer using Shifted Windows. 2021 IEEE/CVF International Conference on Computer Vision (ICCV), 9992–10002. https://doi.org/10.1109/iccv48922.2021.00986
- Ren, S., He, K., Girshick, R., Sun, J. (2017). Faster R-CNN: Towards Real-Time Object Detection with Region Proposal Networks. IEEE Transactions on Pattern Analysis and Machine Intelligence, 39 (6), 1137–1149. https://doi.org/10.1109/tpami.2016.2577031
- LLiu, W., Anguelov, D., Erhan, D., Szegedy, C., Reed, S., Fu, C.-Y., Berg, A. C. (2016). SSD: Single Shot MultiBox Detector. Computer Vision – ECCV 2016, 21–37. https://doi.org/10.1007/978-3-319-46448-0_2
- Ba, J. L., Kiros, J. R., Hinton, G. E. (2016). Layer normalization. arXiv. https://doi.org/10.48550/arXiv.1607.06450
- Loshchilov, I., Hutter, F. (2019). Decoupled weight decay regularization. arXiv. https://doi.org/10.48550/arXiv.1711.05101
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2024 Aigerim Bolshibayeva, Sabina Rakhmetulayeva, Baubek Ukibassov, Zhandos Zhanabekov

This work is licensed under a Creative Commons Attribution 4.0 International License.
The consolidation and conditions for the transfer of copyright (identification of authorship) is carried out in the License Agreement. In particular, the authors reserve the right to the authorship of their manuscript and transfer the first publication of this work to the journal under the terms of the Creative Commons CC BY license. At the same time, they have the right to conclude on their own additional agreements concerning the non-exclusive distribution of the work in the form in which it was published by this journal, but provided that the link to the first publication of the article in this journal is preserved.
A license agreement is a document in which the author warrants that he/she owns all copyright for the work (manuscript, article, etc.).
The authors, signing the License Agreement with TECHNOLOGY CENTER PC, have all rights to the further use of their work, provided that they link to our edition in which the work was published.
According to the terms of the License Agreement, the Publisher TECHNOLOGY CENTER PC does not take away your copyrights and receives permission from the authors to use and dissemination of the publication through the world's scientific resources (own electronic resources, scientometric databases, repositories, libraries, etc.).
In the absence of a signed License Agreement or in the absence of this agreement of identifiers allowing to identify the identity of the author, the editors have no right to work with the manuscript.
It is important to remember that there is another type of agreement between authors and publishers – when copyright is transferred from the authors to the publisher. In this case, the authors lose ownership of their work and may not use it in any way.








