Hybrid imputation of biomedical data by using transformers and autoencoders for assessing human biological age
DOI:
https://doi.org/10.15587/1729-4061.2025.340325Keywords:
data imputation, composite architecture, deep learning, functional age, PhenoAge, NHANESAbstract
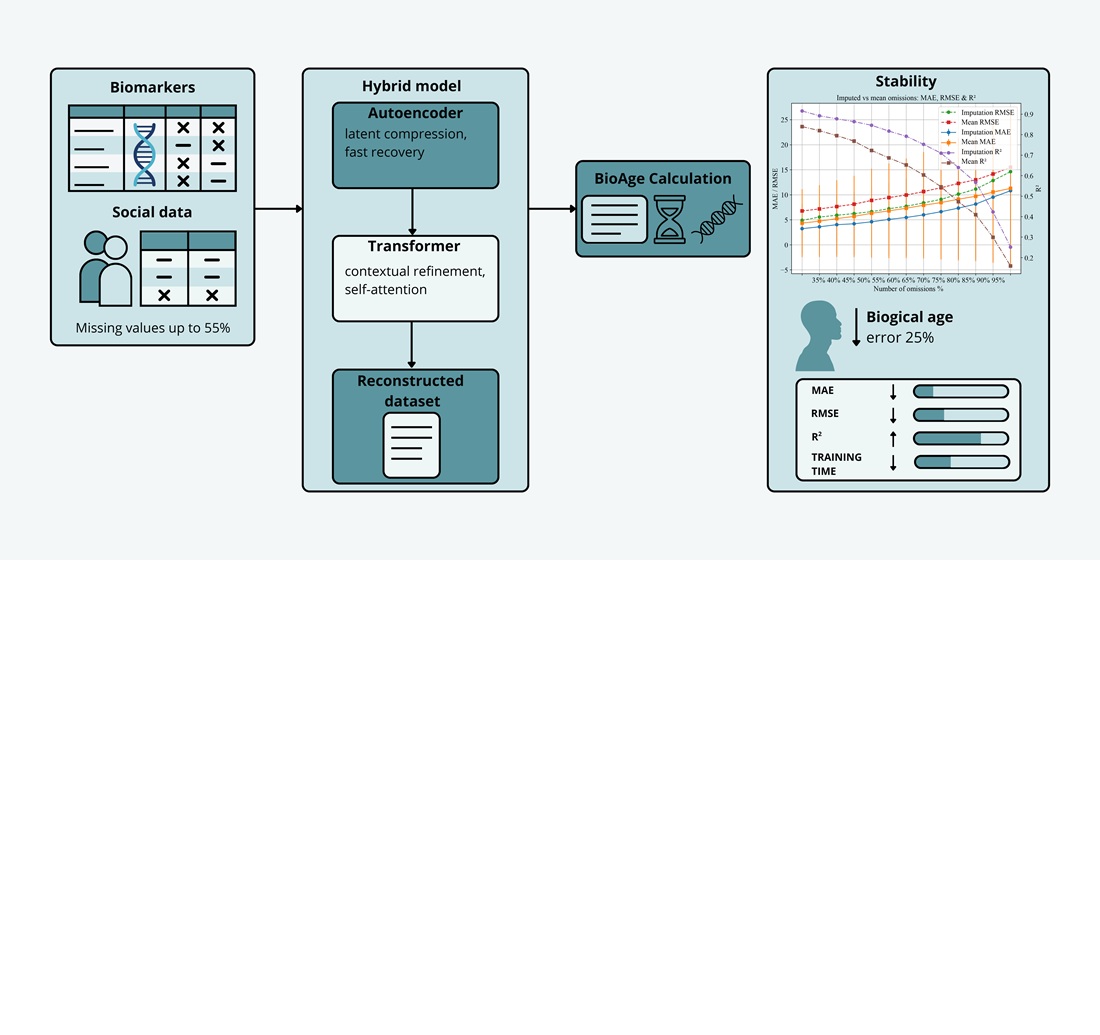
This study investigates the process of restoring missing biomedical and social data for human biological age assessment. The principal challenge is the high rate of missing values in datasets, notably NHANES – up to 40%. This complicates accurate health prediction and reduces the effectiveness of preventive interventions.
To address this issue, deep learning methods, specifically autoencoders and transformers, were employed. The autoencoder provided fast imputation (37.4 s, MAE = 7.54) but less accuracy. The transformer achieved the highest accuracy (246.3 s, MAE = 1.10) yet required substantial resources and showed overfitting risks.
A hybrid architecture has been proposed to combine the advantages of both approaches. On the NHANES dataset (55,081 records and 84 biomarkers), the model demonstrated an optimal balance (54.2 s, MAE = 5.26) and stability with up to 50% missing data. Compared to mean-value imputation, the accuracy of biological age estimation improved by 25%. The coefficient of determination reached 0.9875, and root mean squared error was 35.9, confirming strong consistency of the restored values. Sensitivity analysis revealed stable accuracy up to 55% missing data, after which degradation occurred.
A unique feature of the hybrid approach is the combination of high accuracy with moderate computational cost. This makes the model suitable for medical information systems with incomplete datasets. Practical applications include preventive medicine, biological aging monitoring, and risk group identification.
In the Ukrainian context, the model could enhance biomedical research and digital healthcare while also serving as a foundation for bioinformatics and life expectancy studies
References
- Poliahushko, L., Volkov, O. (2024). Socioeconomic influence on biological age: an overview of current studies and role of artificial intelligence. Telecommunication and information technologies, 3 (84), 120–130. https://doi.org/10.31673/2412-4338.2024.03041234
- Lau, D. T., Ahluwalia, N., Fryar, C. D., Kaufman, M., Arispe, I. E., Paulose-Ram, R. (2023). Data Related to Social Determinants of Health Captured in the National Health and Nutrition Examination Survey. American Journal of Public Health, 113 (12), 1290–1295. https://doi.org/10.2105/ajph.2023.307490
- Kowsar, I., Rabbani, S. B., Samad, M. D. (2024). Attention-Based Imputation of Missing Values in Electronic Health Records Tabular Data. 2024 IEEE 12th International Conference on Healthcare Informatics (ICHI), 177–182. https://doi.org/10.1109/ichi61247.2024.00030
- Casella, M., Milano, N., Dolce, P., Marocco, D. (2024). Transformers deep learning models for missing data imputation: an application of the ReMasker model on a psychometric scale. Frontiers in Psychology, 15. https://doi.org/10.3389/fpsyg.2024.1449272
- Lim, D. K., Rashid, N. U., Oliva, J. B., Ibrahim, J. G. (2024). Unsupervised Imputation of Non-Ignorably Missing Data Using Importance-Weighted Autoencoders. Statistics in Biopharmaceutical Research, 17 (2), 222–234. https://doi.org/10.1080/19466315.2024.2368787
- Horvath, S. (2013). DNA methylation age of human tissues and cell types. Genome Biology, 14 (10). https://doi.org/10.1186/gb-2013-14-10-r115
- Levine, M. E., Lu, A. T., Quach, A., Chen, B. H., Assimes, T. L., Bandinelli, S. et al. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging, 10 (4), 573–591. https://doi.org/10.18632/aging.101414
- Aracri, F., Bianco, M. G., Quattrone, A., Sarica, A. (2025). Bridging the Gap: Missing Data Imputation Methods and Their Effect on Dementia Classification Performance. Brain Sciences, 15 (6), 639. https://doi.org/10.3390/brainsci15060639
- Altamimi, A., Alarfaj, A. A., Umer, M., Alabdulqader, E. A., Alsubai, S., Kim, T., Ashraf, I. (2024). An automated approach to predict diabetic patients using KNN imputation and effective data mining techniques. BMC Medical Research Methodology, 24 (1). https://doi.org/10.1186/s12874-024-02324-0
- Madley-Dowd, P., Curnow, E., Hughes, R. A., Cornish, R. P., Tilling, K., Heron, J. (2024). Analyses using multiple imputation need to consider missing data in auxiliary variables. American Journal of Epidemiology, 194 (6), 1756–1763. https://doi.org/10.1093/aje/kwae306
- Beaulieu-Jones, B. K., Moore, J. H. (2017). Missing data imputation in the electronic health record using deeply learned autoencoders. Biocomputing 2017, 207–218. https://doi.org/10.1142/9789813207813_0021
- Gondara, L., Wang, K. (2018). MIDA: Multiple Imputation Using Denoising Autoencoders. Advances in Knowledge Discovery and Data Mining, 260–272. https://doi.org/10.1007/978-3-319-93040-4_21
- Li, Y., Rao, S., Solares, J. R. A., Hassaine, A., Ramakrishnan, R., Canoy, D. et al. (2020). BEHRT: Transformer for Electronic Health Records. Scientific Reports, 10 (1). https://doi.org/10.1038/s41598-020-62922-y
- Khan, M. A. (2024). A Comparative Study on Imputation Techniques: Introducing a Transformer Model for Robust and Efficient Handling of Missing EEG Amplitude Data. Bioengineering, 11 (8), 740. https://doi.org/10.3390/bioengineering11080740
- He, S., Grant, P. E., Ou, Y. (2022). Global-Local Transformer for Brain Age Estimation. IEEE Transactions on Medical Imaging, 41 (1), 213–224. https://doi.org/10.1109/tmi.2021.3108910
- Urban, A., Sidorenko, D., Zagirova, D., Kozlova, E., Kalashnikov, A., Pushkov, S. et al. (2023). Precious1GPT: multimodal transformer-based transfer learning for aging clock development and feature importance analysis for aging and age-related disease target discovery. Aging. https://doi.org/10.18632/aging.204788
- Wang, X., Chen, H., Zhang, J., Fan, J. (2024). Generative adversarial learning for missing data imputation. Neural Computing and Applications, 37 (3), 1403–1416. https://doi.org/10.1007/s00521-024-10652-x
- Hong, S., Lynn, H. S. (2020). Accuracy of random-forest-based imputation of missing data in the presence of non-normality, non-linearity, and interaction. BMC Medical Research Methodology, 20 (1). https://doi.org/10.1186/s12874-020-01080-1
- Zhou, Y.-H., Saghapour, E. (2021). ImputEHR: A Visualization Tool of Imputation for the Prediction of Biomedical Data. Frontiers in Genetics, 12. https://doi.org/10.3389/fgene.2021.691274
- Bae, C.-Y., Im, Y., Lee, J., Park, C.-S., Kim, M., Kwon, H. et al. (2021). Comparison of Biological Age Prediction Models Using Clinical Biomarkers Commonly Measured in Clinical Practice Settings: AI Techniques Vs. Traditional Statistical Methods. Frontiers in Analytical Science, 1. https://doi.org/10.3389/frans.2021.709589
- United States Department of Health and Human Services. Centers for Disease Control and Prevention. National Center for Health Statistics. National Health and Nutrition Examination Survey (NHANES), 1999-2000 (2012). Inter-university Consortium for Political and Social Research [distributor]. https://doi.org/10.3886/icpsr25501.v4
- Mack, C., Su, Z., Weistreich, D. (2018). Managing Missing Data in Patient Registries. Agency for Healthcare Research and Quality (AHRQ). https://doi.org/10.23970/ahrqregistriesmissingdata
- Chicco, D., Warrens, M. J., Jurman, G. (2021). The coefficient of determination R-squared is more informative than SMAPE, MAE, MAPE, MSE and RMSE in regression analysis evaluation. PeerJ Computer Science, 7, e623. https://doi.org/10.7717/peerj-cs.623
- da Silva, I. N., Hernane Spatti, D., Andrade Flauzino, R., Liboni, L. H. B., dos Reis Alves, S. F. (2016). Multilayer Perceptron Networks. Artificial Neural Networks, 55–115. https://doi.org/10.1007/978-3-319-43162-8_5
- Jinbo, Z., Yufu, L., Haitao, M. (2025). Handling missing data of using the XGBoost-based multiple imputation by chained equations regression method. Frontiers in Artificial Intelligence, 8. https://doi.org/10.3389/frai.2025.1553220
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Volodymyr Slipchenko, Liubov Poliahushko, Oleksandr Volkov, Vladyslav Shatylo

This work is licensed under a Creative Commons Attribution 4.0 International License.
The consolidation and conditions for the transfer of copyright (identification of authorship) is carried out in the License Agreement. In particular, the authors reserve the right to the authorship of their manuscript and transfer the first publication of this work to the journal under the terms of the Creative Commons CC BY license. At the same time, they have the right to conclude on their own additional agreements concerning the non-exclusive distribution of the work in the form in which it was published by this journal, but provided that the link to the first publication of the article in this journal is preserved.
A license agreement is a document in which the author warrants that he/she owns all copyright for the work (manuscript, article, etc.).
The authors, signing the License Agreement with TECHNOLOGY CENTER PC, have all rights to the further use of their work, provided that they link to our edition in which the work was published.
According to the terms of the License Agreement, the Publisher TECHNOLOGY CENTER PC does not take away your copyrights and receives permission from the authors to use and dissemination of the publication through the world's scientific resources (own electronic resources, scientometric databases, repositories, libraries, etc.).
In the absence of a signed License Agreement or in the absence of this agreement of identifiers allowing to identify the identity of the author, the editors have no right to work with the manuscript.
It is important to remember that there is another type of agreement between authors and publishers – when copyright is transferred from the authors to the publisher. In this case, the authors lose ownership of their work and may not use it in any way.








